Session Information
Date: Monday, November 18, 2024
Title: Systemic Sclerosis & Related Disorders – Basic Science Poster II
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: Systemic sclerosis (SSc) is a chronic, progressive autoimmune disorder with significant morbidity and mortality. The lack of effective therapies for SSc is, in part, due to the poor understanding of the molecular and cellular mechanisms underlying fibrosis pathogenesis. To deepen our understanding of disease progression, we interrogated published transcriptomics datasets to deconvolute molecular signatures of SSc over time. The integrated gene signatures were applied to preclinical models of SSc, enabling optimization of models based upon human disease-relevant endpoints. Antifibrotic agents, including a tool LPAR1 inhibitor, were used for model validation.
Methods: Transcriptomics profiling and enrichment analyses were used to integrate three published datasets (GSE130955, GSE58095, and GSE181549). We constructed novel biomarker maps that model the dysregulation seen in early SSc (mean duration 1.3yr), late SSc (mean duration 7.7yr), and common to both early and late disease. Select biomarkers from these maps were measured in preclinical models. NHDFs (normal human dermal fibroblasts) and full-thickness human skin explants were injured with complex inflammatory-fibrotic cocktails, in the presence or absence of antifibrotic agents. A 28-day bleomycin-induced mouse model of SSc was evaluated, with and without antifibrotic treatment. Across models, gene expression and secreted proteins were measured.
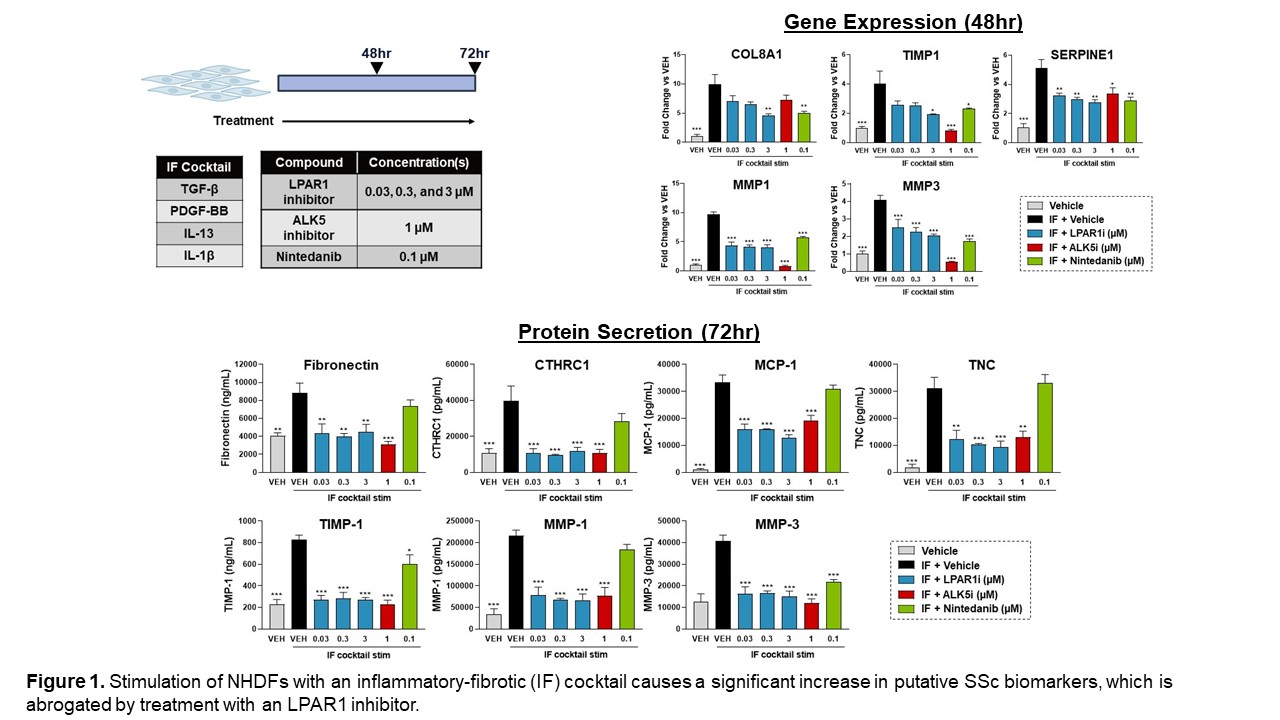
Results: Using published skin transcriptomics datasets, we constructed novel signatures that capture aspects of the pathobiology observed in early and late SSc. We demonstrated that treatment of NHDFs and human skin explants with complex inflammatory-fibrotic cocktails causes an upregulation of multiple genes and proteins identified from these novel SSc signatures, including COL8A1, CTHRC1, MMP-1, and MMP-3. Treatment with a tool LPAR1 inhibitor significantly decreased these endpoints in NHDFs. (Figure 1)
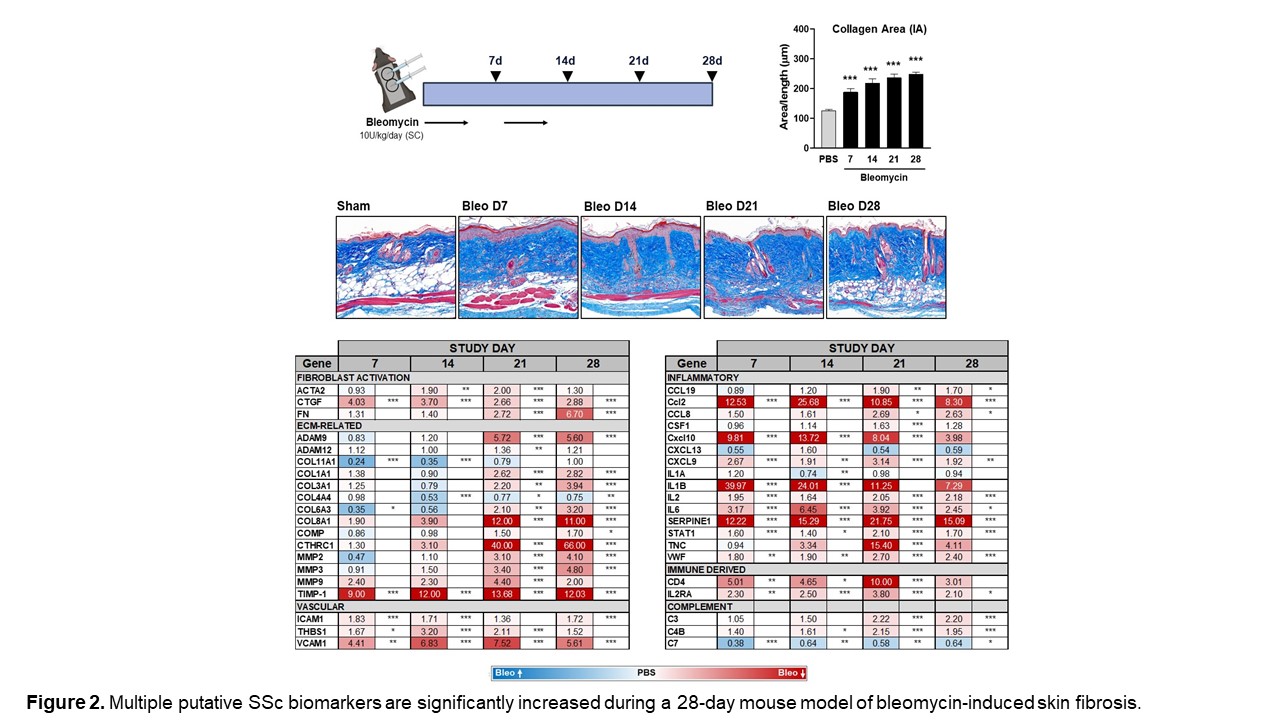
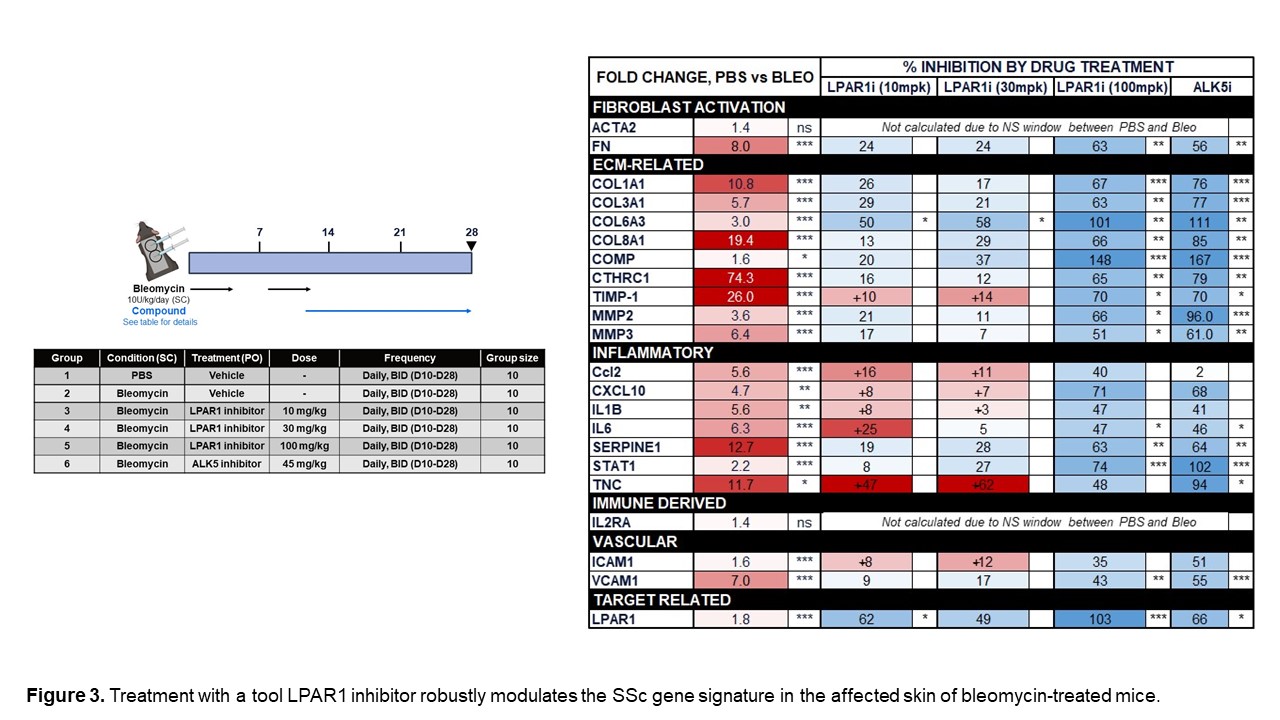
We also assessed the expression of numerous genes in our novel SSc signatures across four time points in a mouse model of bleomycin-induced skin fibrosis to determine whether these changes are recapitulated in this model. Figure 2 highlights genes that were upregulated in both human SSc and the mouse bleo model. Furthermore, we demonstrated that many of the genes that are significantly increased in the skin of bleomycin-treated mice can be significantly decreased by treatment with an LPAR1 inhibitor. (Figure 3) Continuing to identify pathways in the mouse bleomycin model that best reflect changes seen in SSc skin will help improve the translatability of this model.
Conclusion: By profiling three highly-quality SSc skin transcriptomics datasets, we identified key affected pathways and processes throughout SSc pathogenesis. We used these signatures to identify disease-relevant endpoints and guide the optimization of preclinical models to recapitulate key aspects of SSc pathobiology. We also demonstrated that treatment with a tool LPAR1 inhibitor modulates these signatures. These human-centric models will allow us to improve characterization of fibrosis mechanisms and assessment of compound efficacy against key disease processes.
To cite this abstract in AMA style:
Reinke-Breen L, Leys L, Kim S, Olson L, Marinopoulos A, Butler C, Van Camp J, Hazelwood L. Leveraging Novel Systemic Sclerosis Disease Signatures to Build a Humanized Drug Discovery Funnel [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/leveraging-novel-systemic-sclerosis-disease-signatures-to-build-a-humanized-drug-discovery-funnel/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/leveraging-novel-systemic-sclerosis-disease-signatures-to-build-a-humanized-drug-discovery-funnel/