Session Information
Session Type: Abstract Session
Session Time: 3:00PM-4:30PM
Background/Purpose: Psoriasis (Pso) is a chronic immune mediated inflammatory skin disease. Up to 24% of Pso patients develop psoriatic arthritis (PsA). Pathophysiology of PsA is complex and not completely understood. ‘Omics’ studies involve global profiling of biomolecules-genes, RNA, proteins, metabolites using high-throughput technology. Single omics studies in psoriatic disease (PsD) have identified pathways common to both PsA and Pso without arthritis (PsC). Multi-omics integration is a promising strategy that combines different types of omics data to obtain a holistic view of the complex disease pathways. We hypothesize that integrating multiple types of single omic datasets from publicly available independent omics studies comparing PsA and PsC may identify novel PsA-specific pathways.
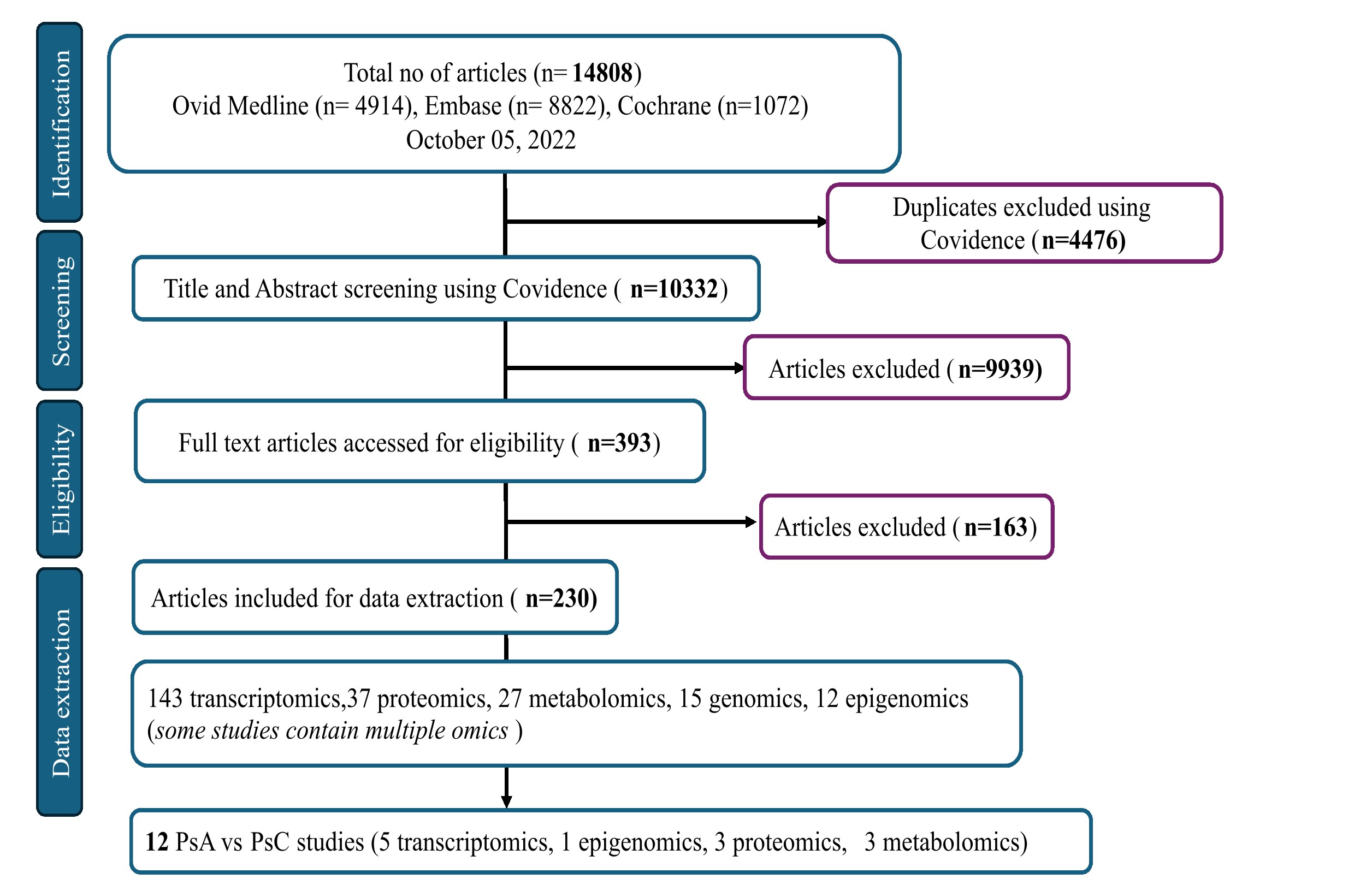
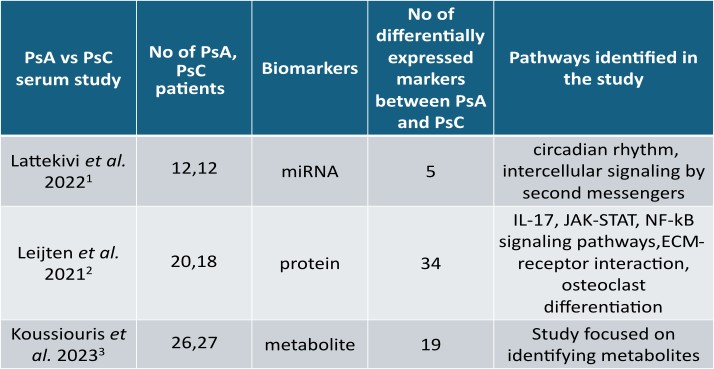
Methods: A scoping review was conducted to curate all publicly available omics studies in the field of PsD from three databases: Ovid Medline, Embase and Cochrane Central. Inclusion criteria comprise all English language studies related to Pso and PsA investigating biomarkers/molecular signatures in human subjects using non-targeted high throughput experiments. Lists of differentially expressed markers, study and clinical information were extracted from all papers that passed the eligibility criteria (Figure 1), to develop a multi-omics data integration portal for PsD (PsDIP). To demonstrate the usability of this portal in identifying PsA-specific pathways, we conducted a preliminary integrative analysis. Lists of differentially expressed proteins, microRNAs (miRNAs) and metabolites from three independent single omics studies comparing serum samples of PsA and PsC patients were collected from PsDIP (Figure 2). All differentially expressed markers were integrated using the following bioinformatics tools: mirDIP (miRNA Data Integration Portal) v5.2, IID (Integrated Interactions Database) ver. 2021-05, STITCH (Search Tool for Interacting Chemicals) v5, pathDIP (Pathway Data Integration Portal) v5, and NAViGaTOR (Network Analysis, Visualization, & Graphing TORonto) v3. Single omics markers (proteins, metabolites and miRNAs) were connected via a network of biological interactions and overlapping pathways.
Results: 5 miRNAs, 34 proteins and 19 metabolites differentially expressed between PsA and PsC were derived from the three independent studies selected. 71 target genes of the 5 miRNAs were found to be connected with 10 proteins and 19 gene interactors of 3 metabolites via protein-protein interactions and 71 statistically significant pathways (q< 0.05) were found to be common among them. 39 are found to be relevant to PsA from literature. 25 of the 39 pathways are potentially important pathways for PsA not identified by the PsA single omics studies in PsDIP such as RANKL, oncostatin M, HIF-1, PDGFR-beta, M-CSF, IL-7, and IL-18 signaling pathways (Figure 3).
Conclusion: Multi-omics integration of independent single omics serum datasets identified key pathways related to osteoclastogenesis, angiogenesis and inflammation which are important to PsA pathophysiology. Further analyses and validation are ongoing.
References:
1. Lättekivi, F.; Guljavina, I.; Midekessa, G.; Viil, J.; Heath, P. R.; Bæk, R.; Jørgensen, M. M.; Andronowska, A.; Kingo, K.; Fazeli, A. Profiling Blood Serum Extracellular Vesicles in Plaque Psoriasis and Psoriatic Arthritis Patients Reveals Potential Disease Biomarkers. Int J Mol Sci 2022
2. Leijten, E.; Tao, W.; Pouw, J.; van Kempen, T.; Olde Nordkamp, M.; Balak, D.; Tekstra, J.; Muñoz-Elías, E.; DePrimo, S.; Drylewicz, J.; et al. Broad proteomic screen reveals shared serum proteomic signature in patients with psoriatic arthritis and psoriasis without arthritis. Rheumatology (Oxford) 2021
3. Koussiouris, J.; Looby, N.; Kulasingam, V.; Chandran, V. A Solid-Phase Microextraction-Liquid Chromatography-Mass Spectrometry Method for Analyzing Serum Lipids in Psoriatic Disease. Metabolites 2023
To cite this abstract in AMA style:
Ghosh S, Pastrello C, Cruz-Correa O, Ganatra D, Oikonomopoulou K, Anderson M, Jurisica I, Chandran V. Identification of Psoriatic Arthritis-related Pathways Using Multi-omics Data Integration [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/identification-of-psoriatic-arthritis-related-pathways-using-multi-omics-data-integration/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/identification-of-psoriatic-arthritis-related-pathways-using-multi-omics-data-integration/