Session Information
Session Type: Poster Session B
Session Time: 10:30AM-12:30PM
Background/Purpose: As genes constitute < 2% of our genomes, there is a need to explore potential roles of other genomic elements in autoimmune disease. We focus on short interspersed elements (SINEs) in this study. They are 260-300 bp elements that occupy roughly 13% of our genomes. There are two major types of SINEs: Alu elements, and mammalian-wide interspersed repeats (MIRs). SINEs have been implicated in multiple biologic functions including gene transcription regulation, alternative splicing, nuclear retention, mRNA decay, and forming double stranded RNA that can trigger interferon signaling. In addition, Alus play a role in binding of NF-κB to the IFN-β enhancer.
Methods: We aligned muscle biopsy RNA-sequencing data (n=152) to the T2T reference genome, which is the first truly complete human genome, and quantified the expression of SINEs at the locus level. Analysis was done by myositis group (dermatomyositis [DM]=39, anti-synthetase syndrome [AS]=15, immune-mediated necrotizing myopathy [IMNM]=51, inclusion body myositis [IBM]=15) vs normal control (n=32). After differential expression testing by myositis groups (alpha=0.05), SINEs with unique expression in each group were identified. SINEs can be expressed passively with gene transcription when they’re embedded in exons or introns; or they can be transcribed through their own promotors. Therefore, we classified SINEs based on their genomic coordinates into those that fall inside: exons, introns, promoter regions, or outside these regions (intergenic). Finally, we correlated the top 10 differentially expressed SINEs by group to interferon stimulated genes (ISGs). P values were adjusted for multiple comparisons.
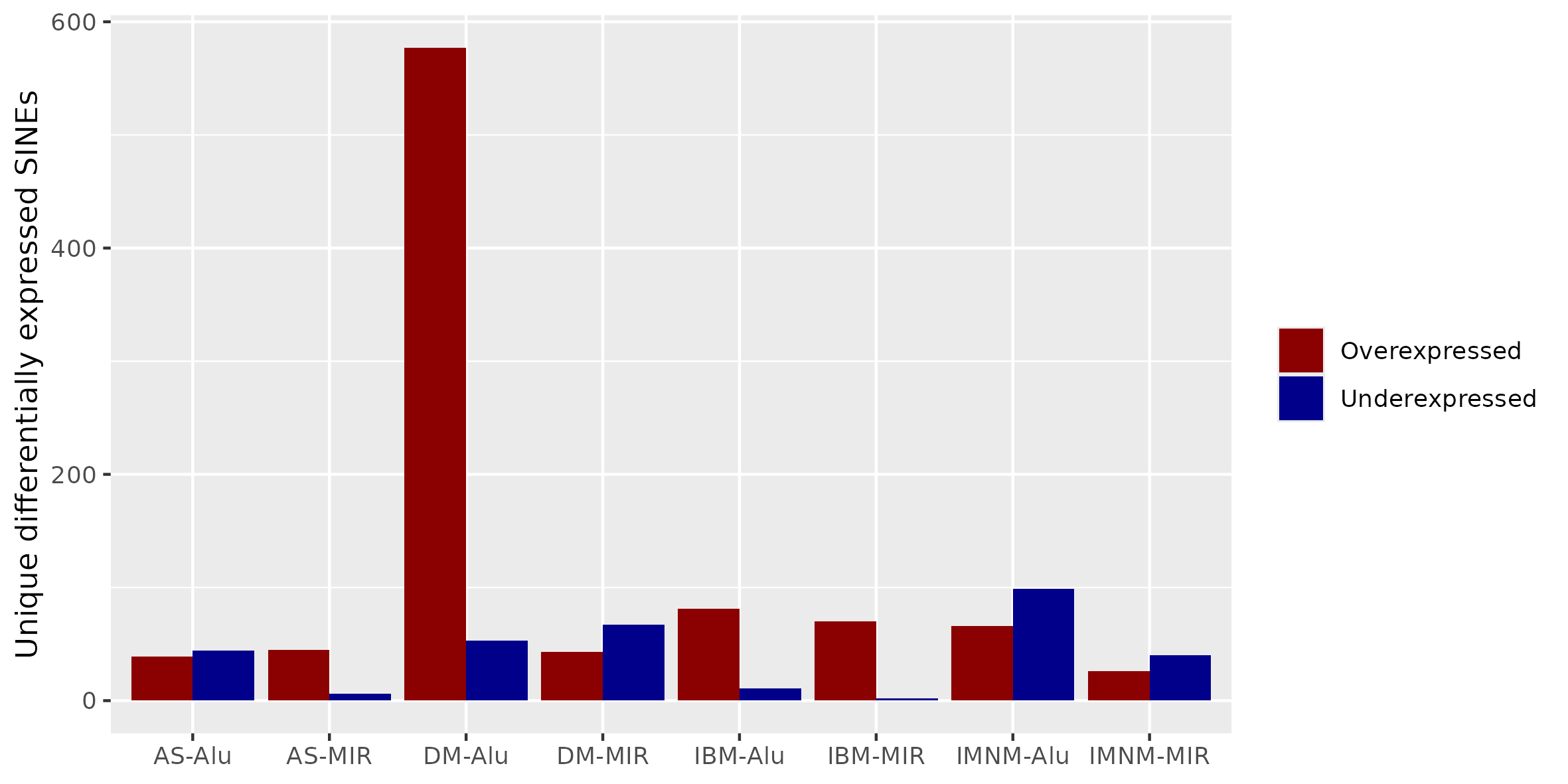
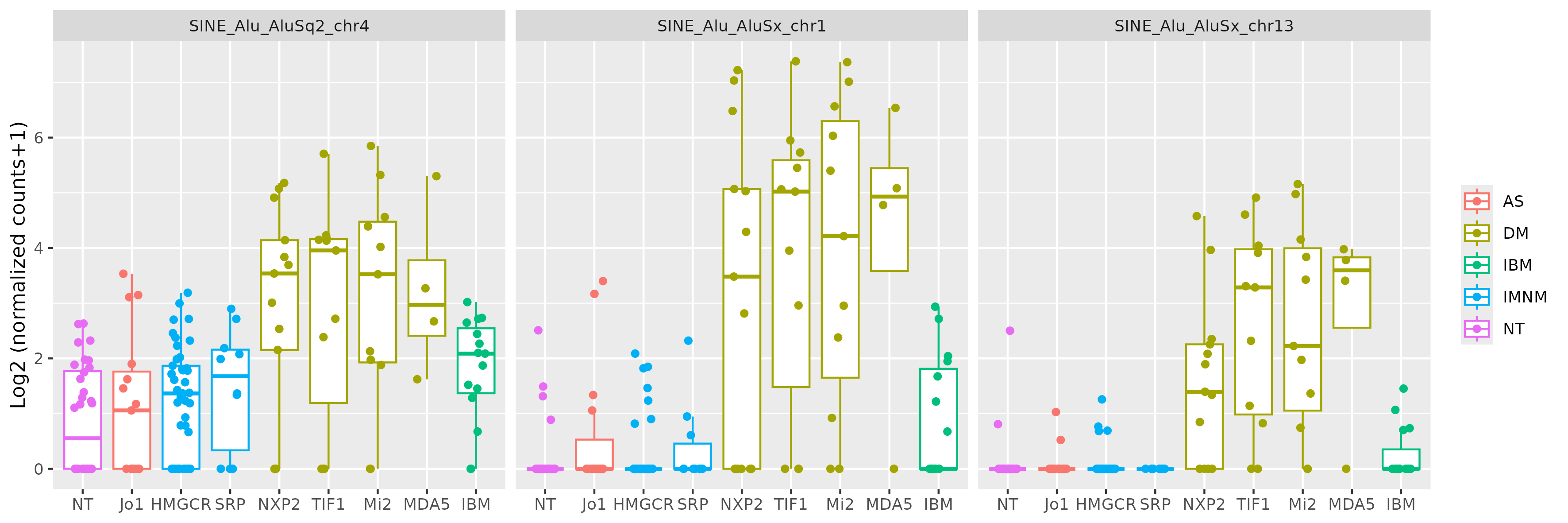
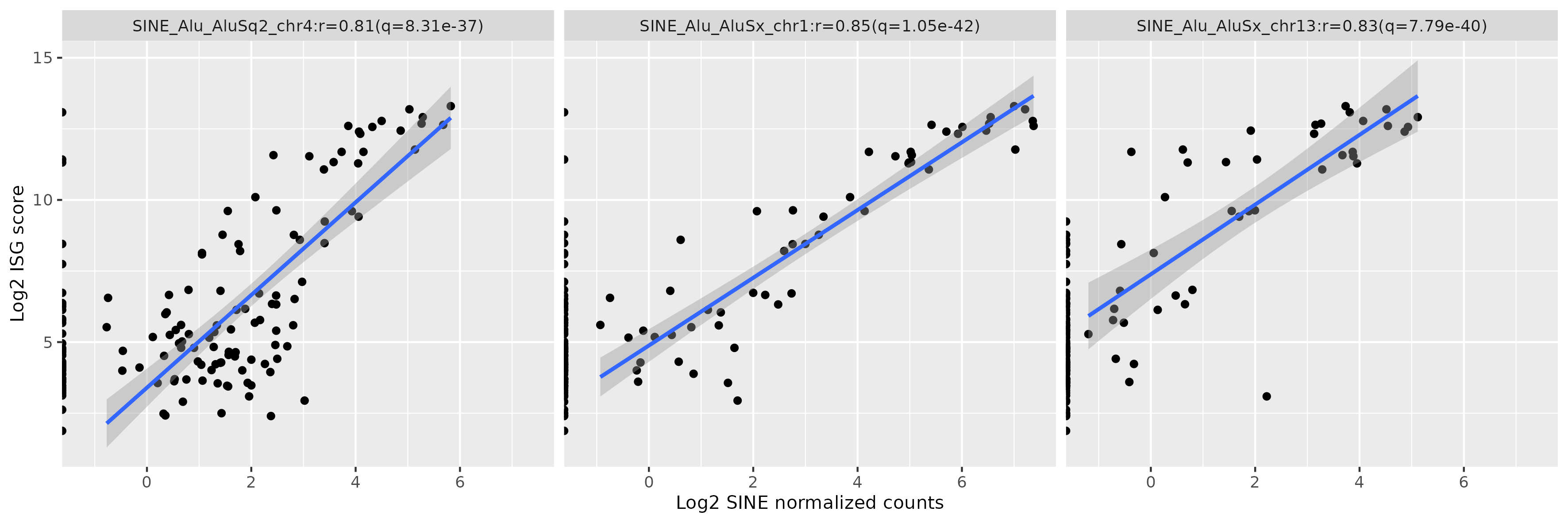
Results: There were many differentially expressed SINEs in each myositis group vs controls (range 1,749-4,568 elements). Uniquely expressed SINEs with ≥2 fold change were highest in DM (740 elements, vs a range of 134-233 in other groups). DM had 620 overexpressed SINES (93% Alu elements), and 120 underexpressed SINEs (56% MIRs), see Figure 1. While IMNM was the only group with more underexpressed (n=140) than overexpressed elements (n=93). In contrast to DM, AS had more MIRs (n=45) than Alus (n=39) that were overexpressed, and mostly underexpressed Alus (n=44), compared with underexpressed MIRs (n=6). Overall, most SINEs were intergenic, including in DM (79%). Of the top 10 differentially expressed Alus in DM, eight were linearly correlated with high ISGs, including three intergenic elements (Figure 2) with strong correlation coefficients (0.81-0.85, q value < 1.0e-36, Figure 3). Similar analyses showed weaker associations of ISGs with top expressed SINEs in AS and IBM, and no significant associations in IMNM.
Conclusion: DM is characterized by unique expression of hundreds of Alu elements that are not seen in other myositis types or in normal muscle. The majority of these Alus were intergenic indicating that they were likely transcribed primarily through their own promotors, and not through passive transcription inside introns and exons of transcribed genes. This analysis reveals a new aspect of the altered biology in dermatomyositis that has not been described before, opening new research avenues to unravel the pathogenesis of myositis.
To cite this abstract in AMA style:
Najjar R, Mammen A, Mustelin T. Intergenic Alu Elements Are Uniquely Expressed in Dermatomyositis and Correlate with Interferon Stimulated Genes [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/intergenic-alu-elements-are-uniquely-expressed-in-dermatomyositis-and-correlate-with-interferon-stimulated-genes/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/intergenic-alu-elements-are-uniquely-expressed-in-dermatomyositis-and-correlate-with-interferon-stimulated-genes/