Session Information
Session Type: Poster Session B
Session Time: 10:30AM-12:30PM
Background/Purpose: Systemic lupus erythematosus (SLE) is a complex genetic disease in which B cells play a critical role in pathogenesis. Previous genome-wide association studies (GWAS) have identified many SLE risk loci and genes important for B cell function. However, it is still unclear how the causal variants within these associations translate to the dysregulated molecular mechanisms that drive the disease. Identifying and characterizing dynamic eQTL effects specifically in primary B cell subtypes treated with an SLE-like inflammatory stimulus using single-cell RNA sequencing (scRNA-seq) may offer new insights into variant-to-function relationships.
Methods: Primary B cells were isolated from 19 healthy donors of Northern European ancestry, then split and cultured for 48 hours with or without a physiologically relevant, defined B cell stimulation to model human SLE: B cell receptor crosslinking IgM/G Fab fragment, CD40 ligand, and TLR7 agonist R837. After 48 hours of culture, the cells were collected and subjected to genotyping and scRNA-seq. The Poisson Mixed Effect (PME) modeling was applied to identify dynamic eQTLs. First, Harmony was used with 10 principal components (PCs) to remove sample- and treatment-specific effects. Each PC was calculated as an independent, continuous, and dynamic cell state covariate. Candidate eQTLs were then fitted with the PME model to test interactions of genotype effects with multiple covariates (donor fixed effects: age, sex, and genotype PCs; cell fixed effects: gene expression PCs, percent mitochondrial UMIs, and number of UMIs; random effects: donor and batch).
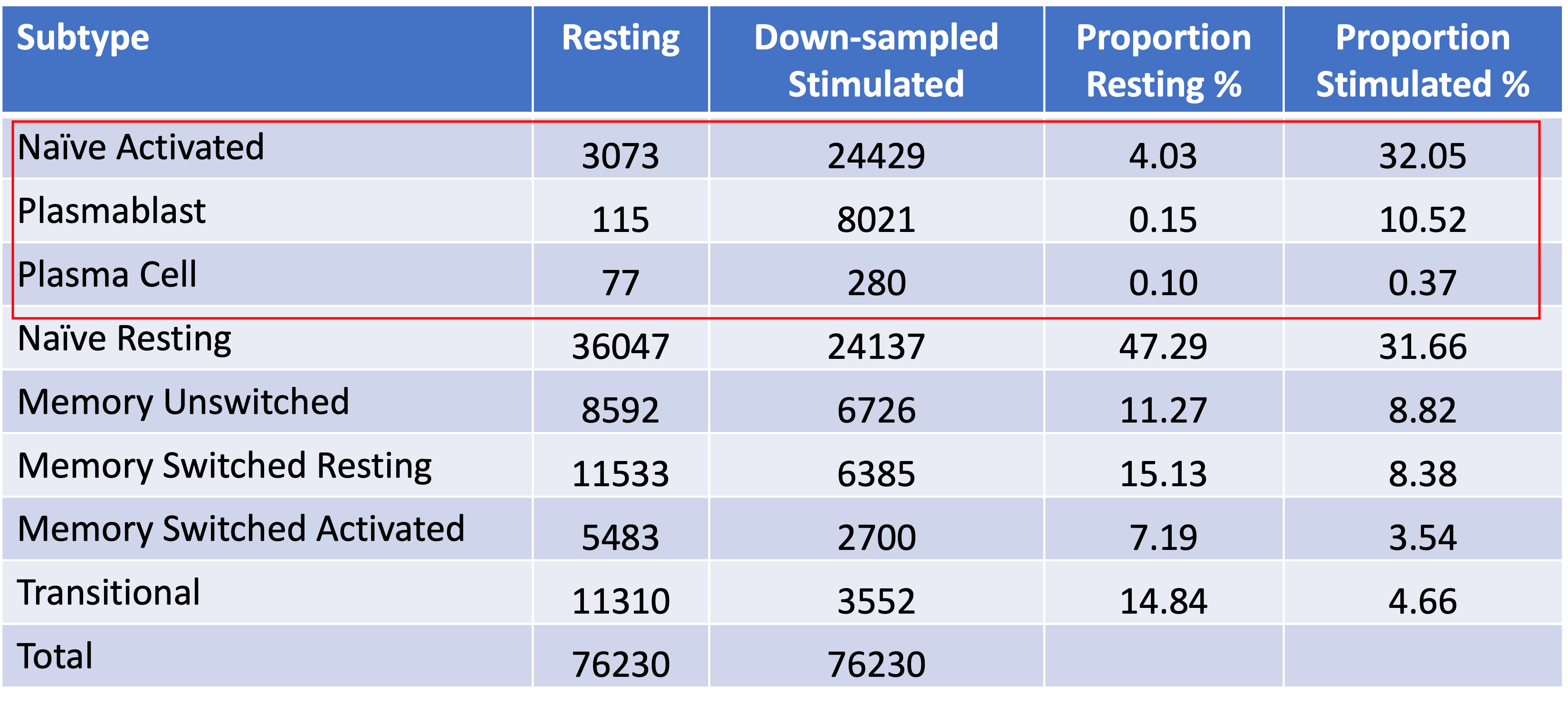
Results: B cell size and viability were significantly increased in response to stimulation. Cell surface CD86 expression also increased, confirming the efficacy of the stimulation. A total of 186,240 B cells (76,230 resting and 110,010 stimulated) were recovered in scRNA-seq. Eight B cell subtypes were manually assigned according to the marker genes of each cluster: transitional cells, naïve resting, naïve activated, memory unswitched, memory switched resting, memory switched activated, plasmablasts, and plasma cells. Three B cell subsets were significantly increased after stimulation: plasmablasts (69.7-fold), naïve activated cells (7.9-fold), and plasma cells (3.6-fold) (Table 1). From an input of 1,353 most significant eQTLs identified in our previous bulk RNA-seq data, we identified 254 eQTLs that demonstrated a significant interaction between genotype and cell state (likelihood ratio-test, FDR < 0.05). For example, two BLK eQTL variants positioned on the same haplotype (rs1478898 and rs2061830) demonstrated dynamic eQTL effects across B cell subsets but opposing responses to stimulation.
Conclusion: scRNA-seq revealed B cell subtype-specific dynamic eQTL effects in response to stimulation. The PME analysis was able to identify dynamic eQTLs, whose genotype effects are dependent on continuous cell states, and characterize the unique genotype effects at single-cell resolution without the limitations of common pseudobulk approaches. We are currently expanding this dataset with 40 more samples to increase power.
To cite this abstract in AMA style:
Fu Y, Murphy D, Pelikan R, Kelly J, Grundahl K, Pasula S, Tessneer K, Gaffney P. Dynamic eQTLs from Activated Human B Cells Suggest Cell-state Dependent Effects for GWAS Risk Variants in BLK [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/dynamic-eqtls-from-activated-human-b-cells-suggest-cell-state-dependent-effects-for-gwas-risk-variants-in-blk/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/dynamic-eqtls-from-activated-human-b-cells-suggest-cell-state-dependent-effects-for-gwas-risk-variants-in-blk/