Session Information
Session Type: Poster Session B
Session Time: 9:00AM-11:00AM
Background/Purpose: Recently mitochondria have been recognized as a key player in the pathogenesis of systemic lupus erythematosus (SLE). Given that N6-methyladenosine (m6A) modifications can regulate almost all aspects of RNA metabolism, such as splicing, stability, structure and translation, and are directly related to T cell function, in order to clarify the impact of epigenetic regulation on mitochondrial function, in this study, we focused on m6A-associatedmitochondrial abnormalities and their effects on lupus CD4+ T cells.
Methods: MethylatedRNAimmunoprecipitation sequencing (MeRIP-seq) was performed using peripheral blood mononuclear cells (PBMCs) from SLE patients and healthy controls to screen for mRNA with abnormal m6A modification and differential expression. The expression of human NADH-ubiquinone oxidoreductase chain 6(MT-ND6)in PBMCs from SLE patients and healthy controls was verified by quantitative real-time polymerase chain reaction (qPCR) and western blot, and analyzed for the correlation with clinical variables. The effects of MT-ND6 on T cell mitochondrial function were clarified by detecting reactive oxide species, mitochondrial reactive oxide species, mitochondrial membrane potential (MMP) and adenosine triphosphate (ATP). To observe the effects of MT-ND6 silencing on CD4+ T cells, small interfering RNA (siRNA) was designed and added into in vitro cultures.
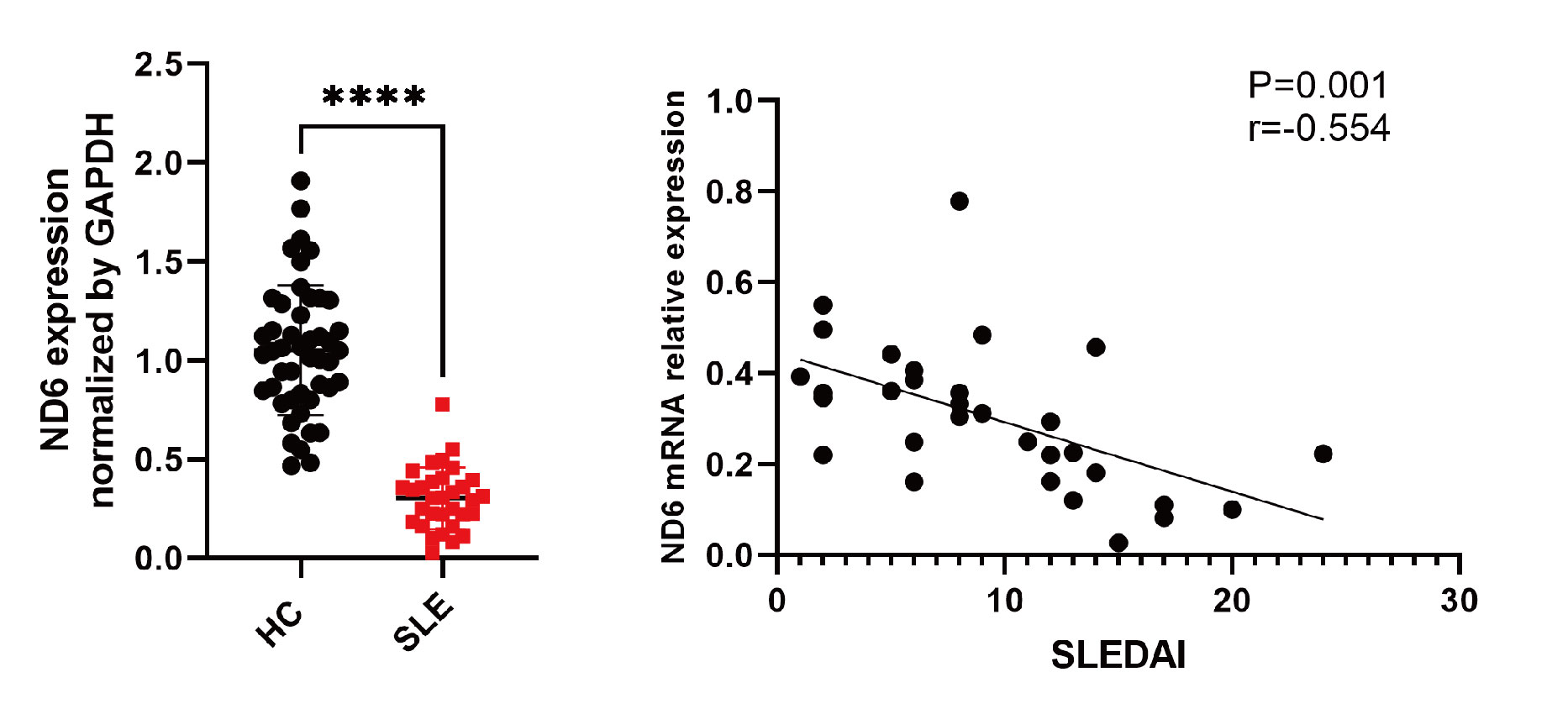
Results: MeRIP-seq identified 370 differentially expressed genes with increased or decreased m6A methylation in lupus, including mitochondrial gene MT-ND6. Independent sample validation showed that both mRNA and protein levels of MT-ND6 in PBMCs from SLE patients were significantly lower than that of healthy controls. MT-ND6 mRNA expression was negatively correlated with SLE disease activity index score (Figure 1) (r=-0.55, p=0.001) and 24-hour urine protein level (r=-0.57, p< 0.05), and was lower in patients with positive anti-Sm or anti-dsDNA antibodies. The expression of MT-ND6 in CD4+ T cells from lupus patients was also significantly decreased compared to healthy controls and was accompanied by significantly increased ROS and mtROS levels, decreased mitochondrial membrane potential and insufficient ATP production. Consistently, when silencing MT-ND6 in CD4+ T cells, ROS and mtROS were significantly increased, while mitochondrial membrane potential and ATP production were reduced.After gene silencing, the levels of IFN-γ and T-bet in CD4+ T cells were significantly increased, and the levels of key inflammatory pathway factors, including STAT4, AP-1 and NF-κB, were all increased.
Conclusion: Low expression of MT-ND6 in lupus can lead to mitochondrial dysfunction, thereby activating CD4+T cells. Our result may provide new therapeutic targets for the treatment of autoimmune diseases.
To cite this abstract in AMA style:
Abdukiyum M, Zhao N, Zheng Y, Alim T, Tang X, Feng X. Down-Regulation of Human NADH-Ubiquinone Oxidoreductase Chain 6 by N6-Methyladenosine Methylation Is Associated with Lupus CD4+ T Cell Activation [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/down-regulation-of-human-nadh-ubiquinone-oxidoreductase-chain-6-by-n6-methyladenosine-methylation-is-associated-with-lupus-cd4-t-cell-activation/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/down-regulation-of-human-nadh-ubiquinone-oxidoreductase-chain-6-by-n6-methyladenosine-methylation-is-associated-with-lupus-cd4-t-cell-activation/