Session Information
Date: Monday, November 13, 2023
Title: (0859–0885) Osteoarthritis & Joint Biology – Basic Science Poster
Session Type: Poster Session B
Session Time: 9:00AM-11:00AM
Background/Purpose: RNA-binding proteins (RBPs) have diverse and essential biological functions, but their role in cartilage health and disease is largely unknown. The objectives of this study were (i) map the global landscape of RBPs expressed and enriched in heathy cartilage and dysregulated in osteoarthritis (OA); (ii) prioritize RBPs for their potential role in cartilage and in OA pathogenesis and as therapeutic targets.
Methods: Our published bulk RNA-sequencing (RNA-seq) data of healthy and OA human cartilage, and a census of 1,542 RBPs were utilized to identify RBPs that are expressed in healthy cartilage and differentially expressed (DE) in OA. Next, our comparison of healthy cartilage RNA-seq data to 37 transcriptomes in the Genotype-Tissue Expression (GTEx) database was used to determine RBPs that are enriched in cartilage. Finally, expression of RBPs was analyzed in our single cell RNA-sequencing (scRNA-seq) data from healthy and OA human cartilage.
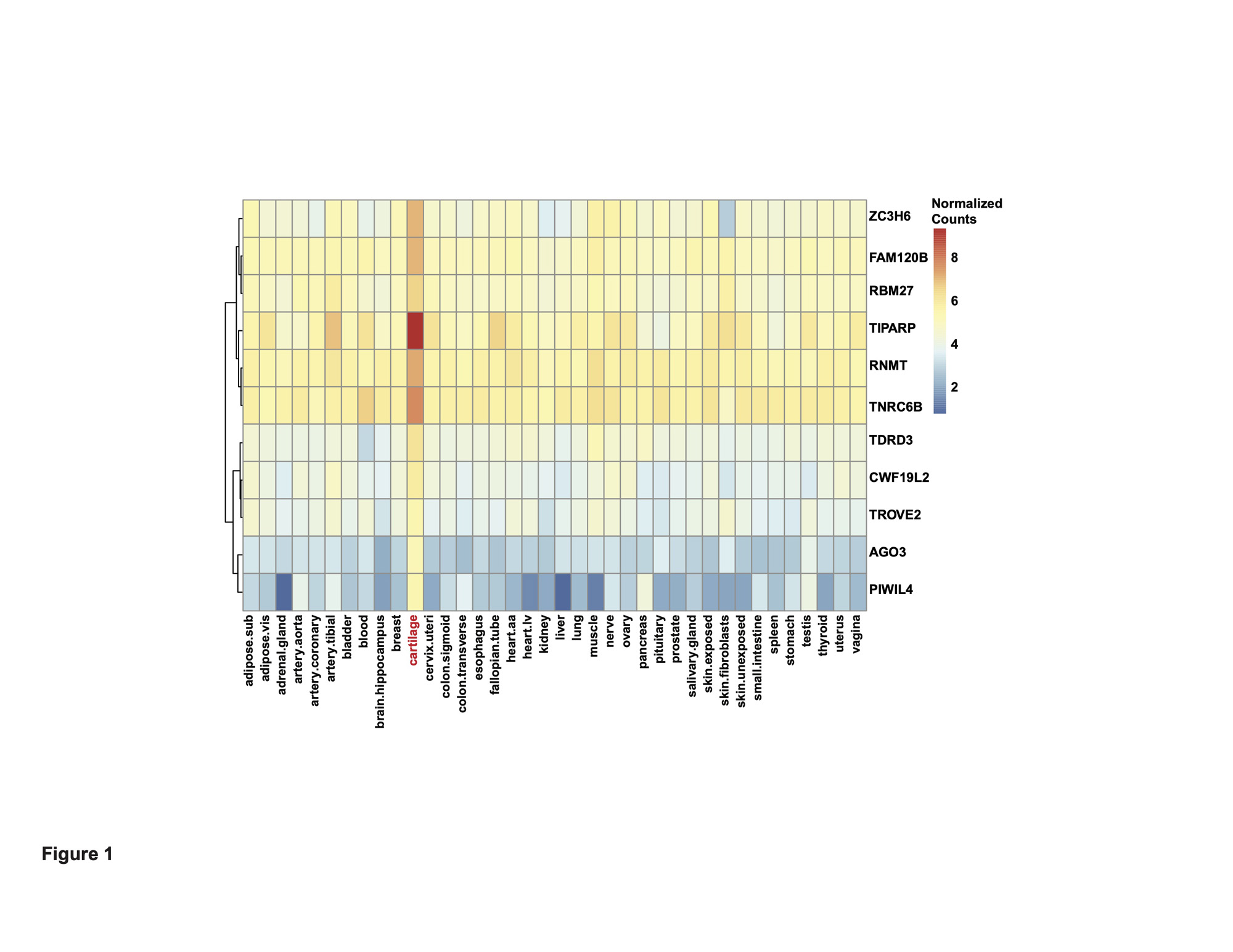
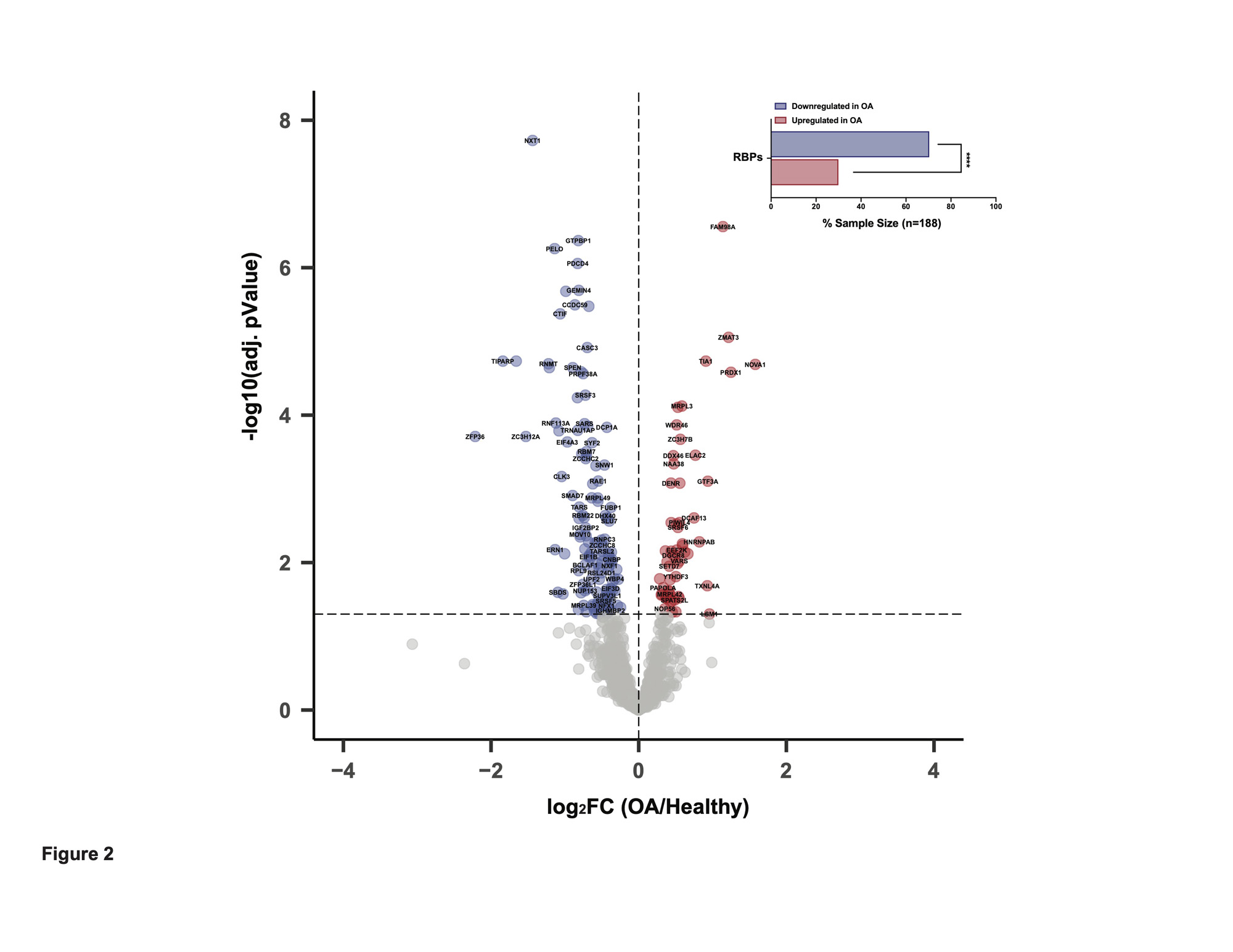
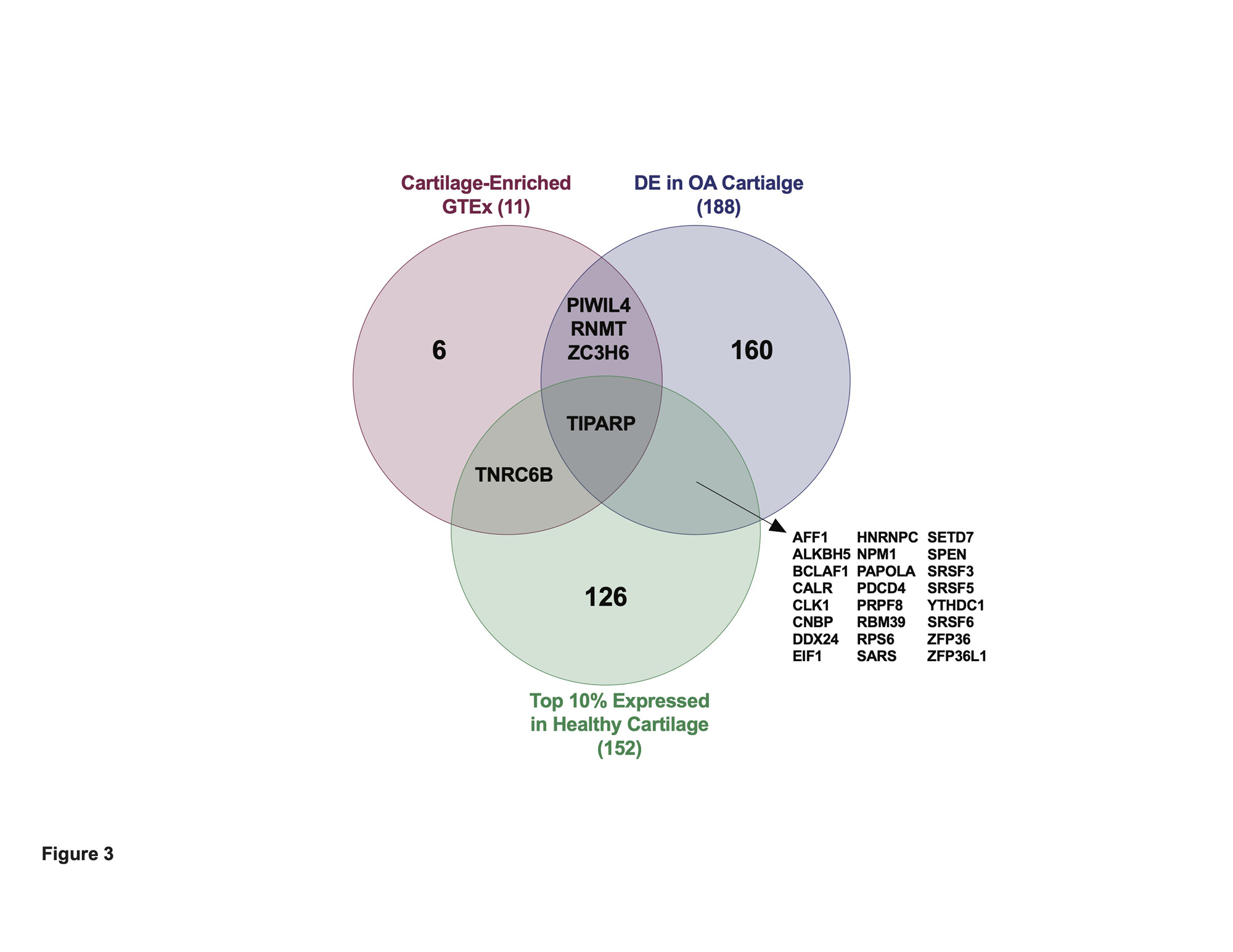
Results: We first investigated in an RNA-seq dataset of healthy human knee cartilage the expression patterns of genes encoding RBPs. Expression of RBPs was higher than nonRBPs, transcription factors (TFs) and long non-coding RNAs (lncRNAs) in healthy cartilage. We next asked which RBPs were specifically enriched in cartilage compared to 37 other issues in GTEx. This analysis revealed 313 cartilage-enriched genes, 11 of which were RBPs (Figure 1). In OA cartilage, 188 RBPs were DE, with a greater proportion downregulated (Figure 2). After identifying the DE RBPs, we investigated their protein associations using STRING. The 188 DE RBPs formed a highly dense protein-protein interaction network. We next investigated both the gene ontology programs and biological pathways associated with the up- and downregulated RBPs. Ribosome biogenesis was enriched in the upregulated RBPs, while splicing and transport were enriched in the downregulated. ChIP-X Enrichment Analysis 3 (ChEA3) was used to identify potential TFs that regulate the DE RBPs. Of the top 20 TFs, the MYC proto-oncogene (MYC) was predicted to regulate the largest number of upregulated RBPs at 42. Of the top 20 TFs, Activating Transcription Factor 1 (ATF1) was predicted to regulate the largest number of downregulated RBPs at 58. To identify a candidate RBP for further studies and as potential therapeutic targets, we investigated other parameters in addition to differential expression including: (i) the top 10% expressed RBPs in healthy cartilage and (ii) those that were cartilage-enriched according to GTEx. Intersecting these three criteria, we identified Tetrachlorodibenzodioxin (TCDD) Inducible Poly(ADP-Ribose) Polymerase (TIPARP) as a candidate RBP (Figure 3). TIPARP was downregulated in OA at both the gene expression and protein levels. scRNA-seq data revealed TIPARP was most significantly downregulated in the ‘pathogenic cluster’ as well as other fibrocytic clusters, suggesting a context-specific effect for TIPARP in chondrocyte function, which was not evident in the bulk RNA-seq data alone.
Conclusion: Our global analyses reveal expression patterns of RBPs in healthy and OA cartilage. We also identified TIPARP and other RBPs as novel mediators in OA pathogenesis and as potential therapeutic targets.
To cite this abstract in AMA style:
Swahn H, Olmer M, Lotz M. RNA-Binding Proteins That Are Highly Expressed and Enriched in Healthy Cartilage but Suppressed in Osteoarthritis [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/rna-binding-proteins-that-are-highly-expressed-and-enriched-in-healthy-cartilage-but-suppressed-in-osteoarthritis/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/rna-binding-proteins-that-are-highly-expressed-and-enriched-in-healthy-cartilage-but-suppressed-in-osteoarthritis/