Session Information
Session Type: Poster Session C
Session Time: 1:00PM-3:00PM
Background/Purpose: In a prior study, we described an alternative method for subphenotyping RA patients by the sequence of biologic DMARDs (bDMARDs) they receive over time. We identified 3 clusters: those prescribed mainly TNF-a inhibitors (TNFi) (“TNFi persisters”), those who start TNFi but switch and predominantly remain on abatacept (“TNFi/abatacept”), and those prescribed multiple bDMARD classes (“multi-bDMARD”). In this study, we hypothesized that genetic risk variants for RA and other inflammatory arthritides (IA) may partially explain which patients remain on TNFi, abatacept, or ultimately trial multiple bDMARDs. Therefore, we examined the association between RA treatment cluster, 191 published IA risk alleles1, and a genetic risk score (GRS) from an RA GWAS.2
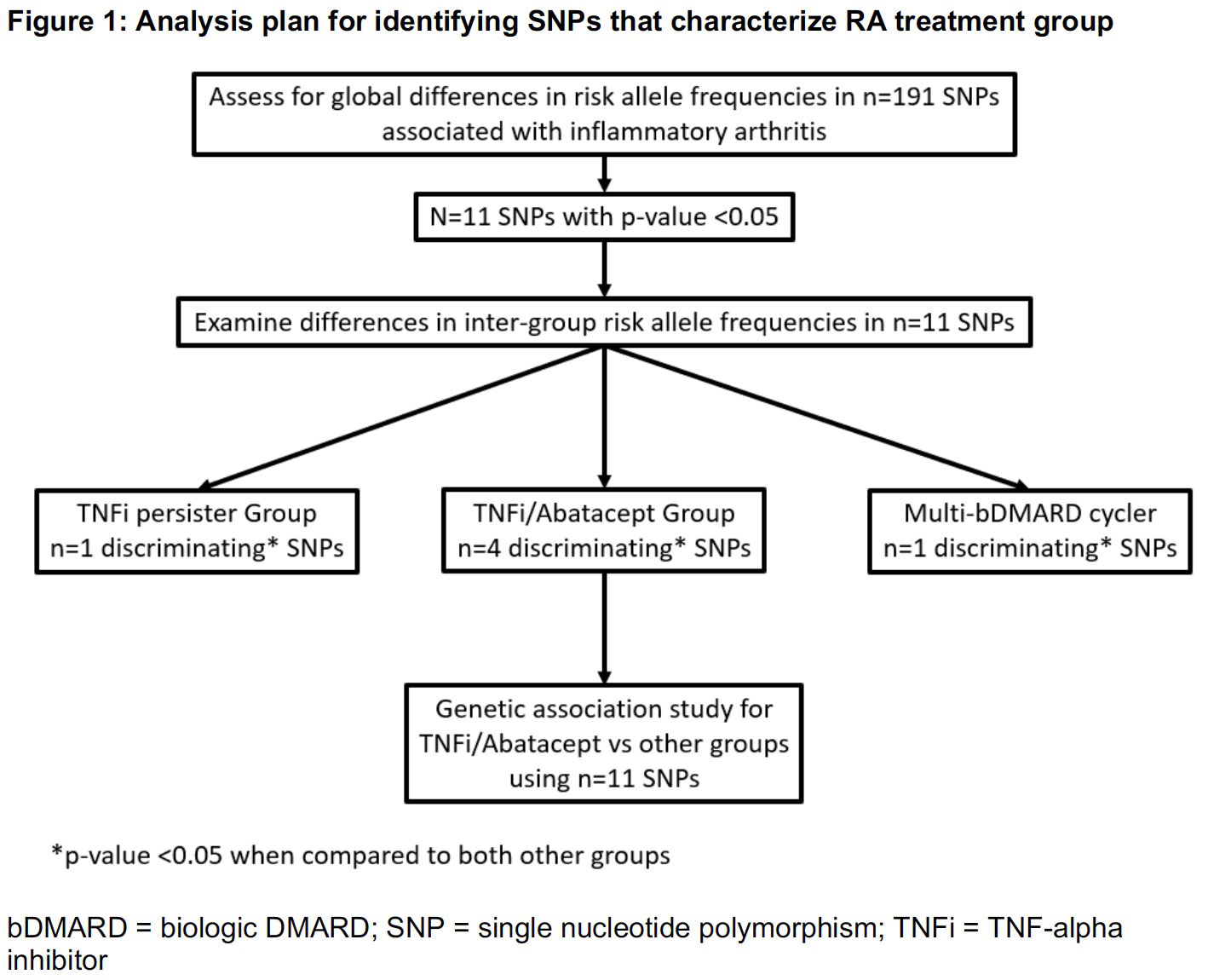
Methods: We studied an electronic health record-based RA cohort linked to genotype data from an institutional biobank of an academic medical center. RA patients who initiated TNFi after 2008 and had >6 months of follow up (mean 5.2 years) were clustered into 1 of 3 treatment groups by prescription history: TNFi persisters, TNFi/abatacept, or multi-bDMARD. We used a stepwise approach to identify the cluster with the greatest difference in allele frequencies compared to the others (Figure 1). First, we tested global differences in allele frequencies in 191 IA-associated single nucleotide polymorphisms (SNPs) across all 3 clusters using chi-squared tests. We then compared inter-group differences for SNPs significant in the global test. Based on the results, our analysis focused on the TNFi/abatacept group. We tested associations of significant SNPs from the global test with the TNFi/abatacept group (vs TNFi persister + multi-bDMARD) using logistic regression adjusted for principal components of ancestry. To test the ability of the SNPs to discriminate among treatment groups, we constructed an unweighted TNFi/abatacept GRS, including SNPs with false discovery rate (FDR) adjusted p< 0.10 in the association testing. We compared receiver operating characteristics of this score to a published RA GRS.
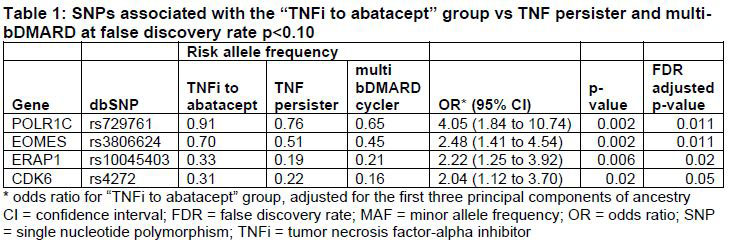
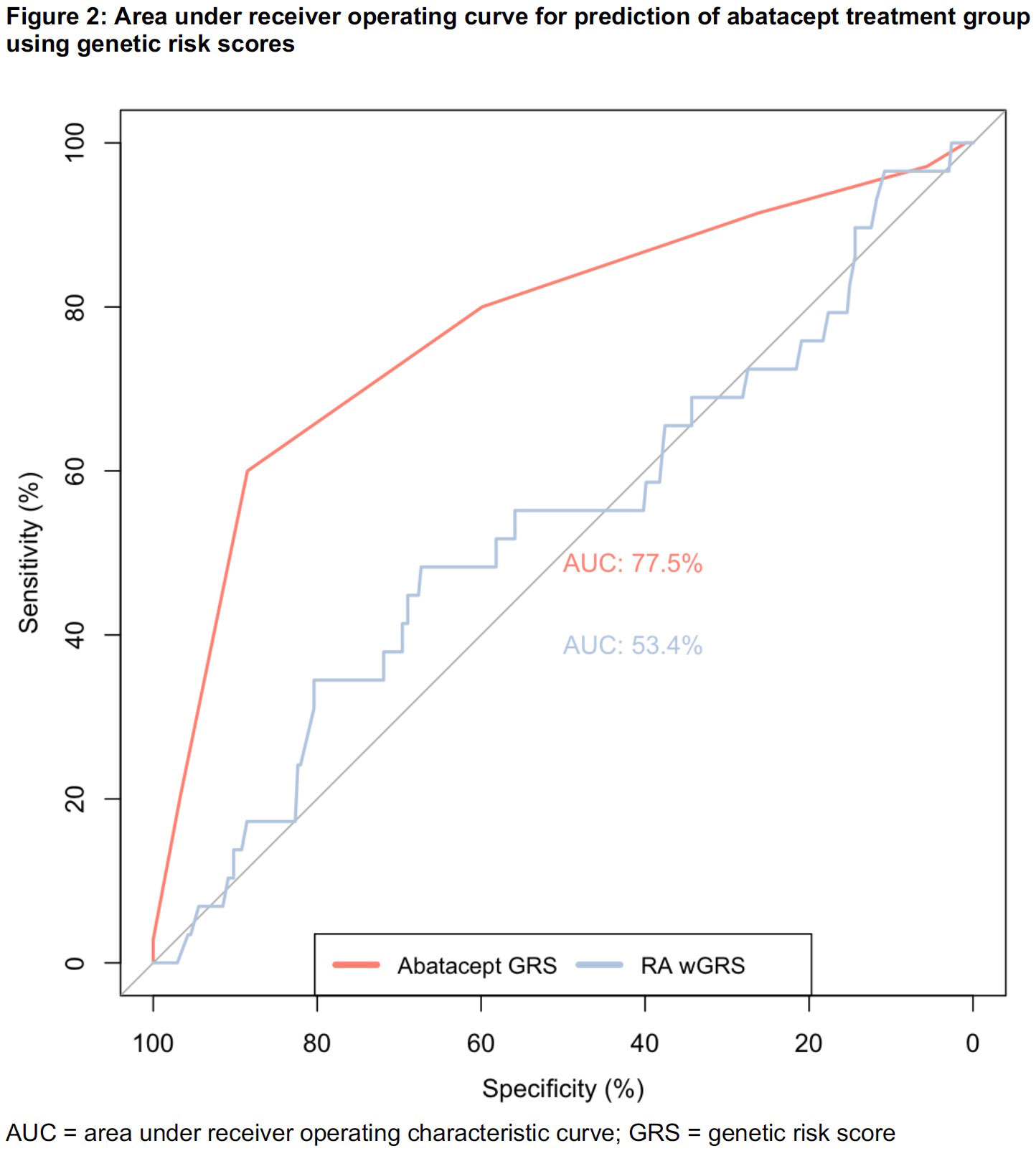
Results: We studied 374 RA patients categorized into the following clusters: TNFi/abatacept (n=35, 9.4%), TNFi persister (n=224, 60%), and multi-bDMARD cycler (n=115, 31%). Based on allele frequencies of the n=191 IA SNPs, the TNFi/abatacept patients had more discriminating SNPs (n=4) at p< 0.05 than the TNFi persisters or multi-bDMARD cycler groups (n=1 each) (Figure 1). The association study adjusted by principal components identified 4 IA SNPs associated with the TNFi/abatacept cluster at FDR adjusted p< 0.10 (Table 1). When we combined these 4 SNPs into a GRS, the area under the ROC curve was 78% for categorizing TNFi/abatacept compared to 53% for the weighted RA GRS (Figure 2).
Conclusion:
Among RA patients clustered by longitudinal bDMARDs prescription data, we observed a difference in genetic variants among subjects who initiate TNFi but switch to and persist on abatacept compared to those persisting on TNFi or using multi-bDMARDs. As overfitting is a limitation of this study, future directions include replication in an independent cohort.1Knevel, Sci Transl Med, 20202Okada, Nature, 2014
To cite this abstract in AMA style:
McDermott G, Cui J, Knevel R, Dahal K, Weisenfeld D, Das P, Karlson E, Cheng S, Raychaudhuri S, Cai T, Liao K. Inflammatory Arthritis Genetic Risk Factors to Predict Treatment Patterns in Rheumatoid Arthritis [abstract]. Arthritis Rheumatol. 2022; 74 (suppl 9). https://acrabstracts.org/abstract/inflammatory-arthritis-genetic-risk-factors-to-predict-treatment-patterns-in-rheumatoid-arthritis/. Accessed .« Back to ACR Convergence 2022
ACR Meeting Abstracts - https://acrabstracts.org/abstract/inflammatory-arthritis-genetic-risk-factors-to-predict-treatment-patterns-in-rheumatoid-arthritis/