Session Information
Date: Sunday, November 13, 2022
Title: Abstracts: Miscellaneous Rheumatic and Inflammatory Diseases II
Session Type: Abstract Session
Session Time: 10:30AM-12:00PM
Background/Purpose: Sarcoidosis is a systemic, non-caseating granulomatous disease driven by a dysregulated immune response to environmental antigens. A wide range of clinical manifestations coupled with unpredictable clinical courses makes studying sarcoidosis challenging. Diagnosis relies on imaging and biopsy and treatment typically includes steroids. Recently, JAK inhibitors have been utilized as steroid-sparing therapies for cutaneous and pulmonary sarcoidosis, however the mechanism is unclear. We present a meta-analysis of 20 clinical transcriptome studies which establishes a shared transcriptional landscape of sarcoidosis, prioritizes candidate biomarkers, explores perturbed pathways and studies the JAK/STAT pathway.
Methods: We searched publicly available transcriptome data repositories for clinical sarcoid datasets with healthy controls and at least 3 biological replicates evaluated with transcriptome analysis and found 20 studies (14 microarray and 6 RNAseq), comprising 316 sarcoid patients and 383 healthy controls. The majority of samples came from peripheral blood; tissue-based samples included lung, skin, anterior orbit, lacrimal gland and lymph nodes. We performed differential gene expression on each of the 20 studies independently with Limma (microarray) and DESeq2 (RNAseq). Results were merged for the 17,705 genes that were evaluated by at least 9 of the studies. For microarray studies, data from the probe with lowest adjusted p-value and largest absolute fold change (FC) was selected. Genes were selected for pathway enrichment analysis if at least 10 studies identified them as differentially expressed. Candidate biomarkers were genes that were consistently differentially expressed in both tissue and peripheral samples with a fold change magnitude of at least 1.5x. Pathway enrichment analysis was performed with Reactome.
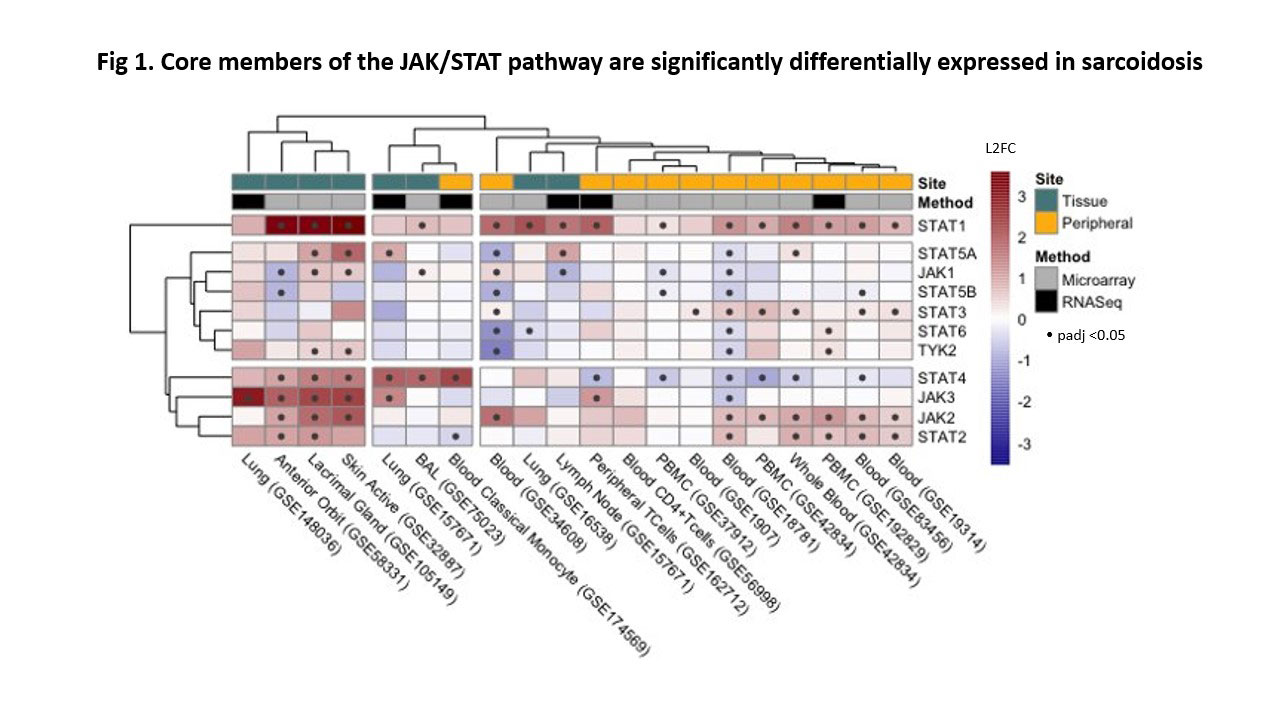
Results: We prioritized 2,349 genes that were differentially expressed in the majority of our datasets. Unsupervised clustering of these studies (FC data) showed a distinct difference between peripheral and tissue sample types. Of the 18 biomarker candidates, some have been associated with sarcoidosis (STAT1, ITGA6, LEF1) and others have been reported in tuberculosis (GBP5, ANKRD22), however, many have not yet been associated with sarcoidosis (RHOH, SPTBN1, UBASH3A). Pathway enrichment identified significant perturbation of interferon signaling (IFNγ, IFNα/β) and antigen presentation which supports two established mechanisms of sarcoid pathophysiology: an abnormal TH1 response and existence of MHC risk alleles. Qualitative exploration of the JAK/STAT pathway (Fig 1) shows a predominant upregulation of the pathway, most strongly in STAT1 and JAK2.
Conclusion: This meta-analysis summarizes the current transcriptional landscape of sarcoidosis, including pathophysiology, biomarkers and therapeutics targeting the JAK/STAT pathway. We suspect JAK2 is an important therapeutic target, by disrupting the JAK/STAT component of TH1 response, and anticipate that there may be therapeutic modulation of other JAKs that attenuate other arms of the adaptive immune system.
To cite this abstract in AMA style:
Lindquist I, Rosenbaum J, Friedman M. Solving Sarcoidosis: A Transcriptome-based Meta-analysis of Clinical Sarcoidosis Studies Illustrates Shared Pathophysiology, Identifies Candidate Biomarkers and Suggests a Therapeutic Mechanism of JAK Inhibition [abstract]. Arthritis Rheumatol. 2022; 74 (suppl 9). https://acrabstracts.org/abstract/solving-sarcoidosis-a-transcriptome-based-meta-analysis-of-clinical-sarcoidosis-studies-illustrates-shared-pathophysiology-identifies-candidate-biomarkers-and-suggests-a-therapeutic-mechanism-of-jak/. Accessed .« Back to ACR Convergence 2022
ACR Meeting Abstracts - https://acrabstracts.org/abstract/solving-sarcoidosis-a-transcriptome-based-meta-analysis-of-clinical-sarcoidosis-studies-illustrates-shared-pathophysiology-identifies-candidate-biomarkers-and-suggests-a-therapeutic-mechanism-of-jak/