Session Information
Session Type: Poster Session B
Session Time: 9:00AM-10:30AM
Background/Purpose: ANCA-associated vasculitis (AAV) is a rare disease associated with substantial morbidity and mortality.1 To enable outcomes and comparative effectiveness studies using large, phenotypically diverse cohorts from big data, a novel case-finding algorithm is needed. Recent studies have assembled cohorts using structured administrative claims data, including billing codes, and ANCA test results but these may lack sensitivity and specificity. In particular, they may exclude important AAV subgroups. Alternatively, using clinical note free-text from the electronic health records to identify features of AAV may improve case-finding. We hypothesized that a machine learning (ML) model can accurately identify AAV cases from clinical notes.
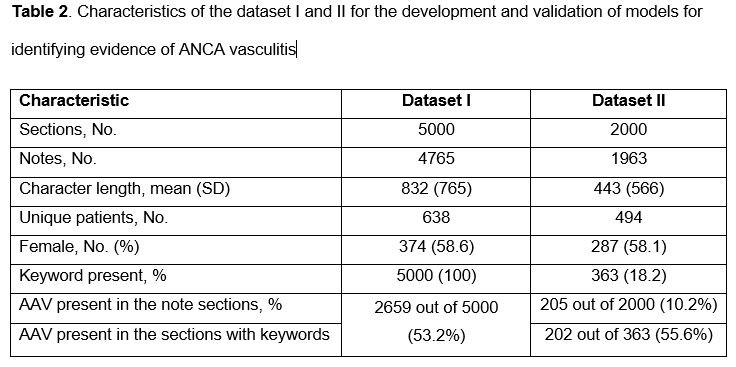
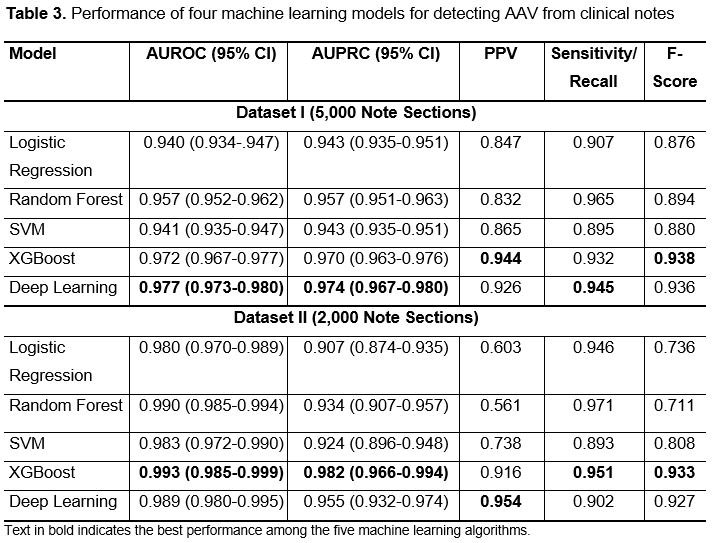
Methods: We used the 2002-2019 Mass General Brigham (MGB) AAV Cohort which is a consecutive inception cohort of PR3- or MPO-ANCA+ AAV patients.2 The notes of the study cohort were extracted from MGB’s enterprise data warehouse and were split into sections using a natural language processing tool. Two reference data sets were created and sections from each were manually labeled as supporting a diagnosis of AAV or not. Data set I contained a random sample of 5000 note sections filtered by a list of keywords potentially related to AAV (Table 1) and was used for model training and testing. Data set II contained 2000 randomly selected sections without keyword filtering to assess whether model performance was dependent on keywords. We compared multiple ML algorithms, including logistic regression, support vector machine (SVM), random forest, XGBoost, and a deep learning algorithm, for classification of note sections for AAV.3 We assessed model performance with positive predictive value (PPV), sensitivity, F-score, area under the ROC curve, and area under the precision and recall curve.
Results: There were 700 patients with notes included. Sections were randomly chosen from 6,728 notes. At least one keyword was present in 18.2% of the note sections from the cohort (Table 2). When restricted to sections that contained selected keywords, more sections contained text supportive of a diagnosis of AAV (increased from 10% to 53-56%). Among the five ML algorithms, XGBoost and deep learning had the best performance in both data sets for identifying sections supportive of a diagnosis of AAV (AUROC 99.3% [95% CI 98.5-99.9%] and 98.9% [95% CI 98.0-99.5%], respectively); there was no significance difference observed between the performance of either approach, but they were superior to logistic regression, random forest, and SVM modeling (Table 3).
Conclusion: ML models, especially XGBoost and deep learning, can accurately classify clinical note sections as reflecting a diagnosis of AAV. Models trained using a data set with keyword filtering were generalizable to a data set without applying keyword filtering. Future studies might apply the models to screen the entire MGB database for AAV cases using clinical notes and compare these approaches to rule-based algorithms.
1. Wallace ZS, et al. Rheumatology (Oxford). 2020;59(9):2308-15.
2. McDermott G, et al. JAMA Intern Med. 2020;180(6):870-6.
3. Wang L, et al. JAMA Netw Open. 2021;4(11):e2135174.
To cite this abstract in AMA style:
Wang L, Laurentiev J, Cook C, Miloslavsky E, Choi H, Zhou L, Wallace Z. Accurate Identification of ANCA-Associated Vasculitis Cases from Clinical Notes Using Machine Learning [abstract]. Arthritis Rheumatol. 2022; 74 (suppl 9). https://acrabstracts.org/abstract/accurate-identification-of-anca-associated-vasculitis-cases-from-clinical-notes-using-machine-learning/. Accessed .« Back to ACR Convergence 2022
ACR Meeting Abstracts - https://acrabstracts.org/abstract/accurate-identification-of-anca-associated-vasculitis-cases-from-clinical-notes-using-machine-learning/