Session Information
Date: Tuesday, November 9, 2021
Title: Metabolic & Crystal Arthropathies – Basic & Clinical Science Poster II (1565–1583)
Session Type: Poster Session D
Session Time: 8:30AM-10:30AM
Background/Purpose: Gout is a complex disease involving changes in urate biology and inflammatory responses, and is associated with comorbidities including metabolic syndrome and cardiorenal disease. Phenome-Wide Association Study (PheWAS) is a novel analytical technique where genetic variants are interrogated against a large set of phenotypic traits. PheWAS are a powerful tool to characterize the clinical significance of genetic variants and gain insights into their biological function.
We performed a PheWAS to define phenotypic traits associated with polygenetic risk of gout using 18 known gout risk loci.
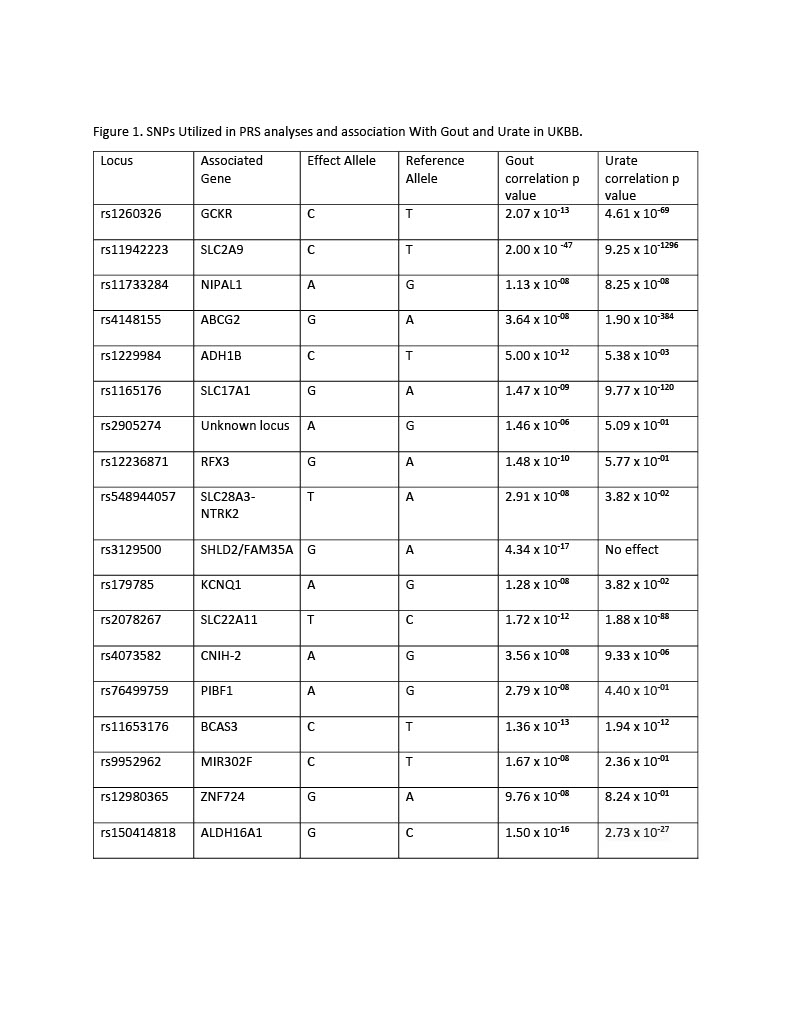
Methods: Gout-associated loci were identified through literature review. Variants in linkage disequilibrium were removed, yielding a total of 18 loci which we analyzed in UK Biobank. The 18 loci were classified as urate-related or non-urate related based on association with serum urate (p< 1 x 10-5). Polygenetic risk scores (PRS) were calculated by summing gout risk increasing alleles weighted by published gout association beta estimates (natural log of odds ratios). PRS were then calculated for all loci, urate-associated loci, and non-urate associated loci, and PheWAS was performed using logistic regression on discrete phenotypic traits (e.g. ICD-10 codes) from the UK Biobank study. To limit ethnic/racial confounders, we performed analyses in unrelated European ancestry UK Biobank study participants (n=382,828).
Results: 10 of the 18 SNPs were urate-associated and 8 were not. The all-gout and urate-associated PRSs revealed phenome-wide significant associations with disorders of iron metabolism (p=7.91 x 10-29 PRS_all, p=9.82 x 10-31 PRS urate), celiac disease (p=1.30 x 10-10 PRS_all, p= 2.42 x 10-12 PRS_urate) and non-celiac disease intestinal malabsorption (p= 1.97 x 10-09 PRS_all, p= 1.86 x 10-10 PRS_urate). The non-urate gout PRS did not identify any phenome-wide significance associations. We determined the urate PRS associations were driven largely by rs1165176 on SLC17A1 (p= 4.2 x 10-81 iron metabolism, p= 5.0 x10-17 celiac disease). Adjustment for HFE SNP rs1800562 (263 kb away on chromosome 6) abrogated the association between rs1165176 and iron metabolism. However, the association of rs1165176 with celiac disease remained significant.
Conclusion: We performed a PheWAS analysis to interrogate the clinical manifestations of polygenetic risk of gout and identified relationships between gout, celiac disease, and intestinal malabsorption. This relationship was driven by genes associated with both gout and hyperuricemia, and not by genes that were associated with gout but not with hyperuricemia. Though epidemiological correlation between gout and ferritin levels was previously described by Fatima et al, our results suggests that linkage between SLC17A1 and HFE may underlie this observation.
To cite this abstract in AMA style:
Stens O, Trang V, Cao S, Terkeltaub R, Salem R. Phenome-Wide Association of Gout Risk Loci [abstract]. Arthritis Rheumatol. 2021; 73 (suppl 9). https://acrabstracts.org/abstract/phenome-wide-association-of-gout-risk-loci/. Accessed .« Back to ACR Convergence 2021
ACR Meeting Abstracts - https://acrabstracts.org/abstract/phenome-wide-association-of-gout-risk-loci/