Session Information
Date: Monday, October 22, 2018
Title: 4M109 ACR Abstract: Systemic Sclerosis & Rel D/Os–Basic Science (1899–1904)
Session Type: ACR Concurrent Abstract Session
Session Time: 4:30PM-6:00PM
Background/Purpose: The molecular mechanisms of systemic sclerosis (SSc) have been difficult to study outside of patient samples. Mouse models often lack key features of the disease, and fibroblast cultures show inconsistent results. We have developed an innovative skin-like tissue of SSc, self-assembled Skin Equivalents (sSE), where we study fibroblast behavior in a 3D microenvironment with epithelial-dermal crosstalk and immune interaction. Here we investigate the molecular changes in SSc sSE and show that it is molecularly similar to SSc skin biopsies.
Methods: Fibroblasts were isolated from SSc patient skin (SScDF) and normal skin (NDF), expanded and seeded into transwell chambers +/- monocytes. SSc patient-derived plasma and healthy control (HC) plasma were incorporated into the culture media during the polarization period. Normal Human Keratinocytes were seeded at 3 weeks for epithelialization, and tissues were harvested after 5 weeks followed by IHC, atomic force microscopy (AFM), RNA-seq, DNA methylation, and ATAC-seq. Cell supernatants were used for multiplex ELISA analysis.
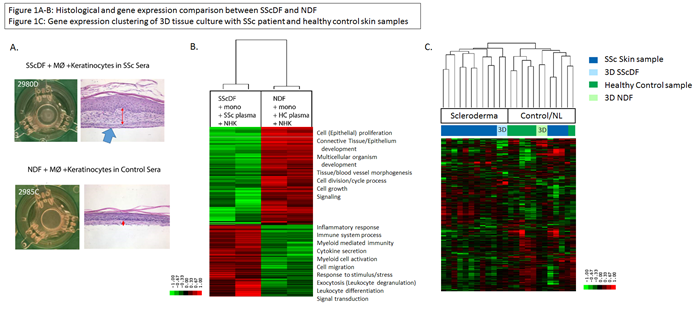
Results: We created 136 samples of sSE from one SScDF line and one NDF line. H&E staining of SScDF sSE showed increased dermal thickness and stiffness compared to NDF sSE (Fig 1A). Differential expression of the SScDF sSE and NDF sSE with autologous plasma and monocytes showed increased expression of genes representing pathways involved in inflammatory/immune system response, myeloid-mediated immunity, myeloid cell activation, and leukocyte differentiation (Fig 1B). The upregulated genes in NDF sSE showed typical pathways involved in epithelial proliferation, tissue morphogenesis, and cell growth. Differential expression of the SScDF sSE +/- monocytes found that sSE tissues with monocytes had increased immune response, immune cell proliferation and activation, macrophage migration, and cell chemotaxis. In samples without monocytes, epithelial cell differentiation and collagen processes were upregulated, but the strong immune signal was missing. Additionally, IL6 and IL13 production increased in SScDF supernatant during tissue development and after the polarization period. Also, analysis of gene expression in SScDF sSE by RNA-seq data demonstrated molecular similarity to human SSc patient skin samples and NDF clustered more closely with HC skin samples (Fig 1C). Lastly, 3D sSE tissues and 2D monolayer fibroblast cultures have distinct DNA methylation patterns.
Conclusion: There is a hierarchy of drivers in the creation of 3D tissues, with the origin of dermal fibroblasts being the biggest modifier of disease morphology and the addition of monocytes being the next biggest factor in developing the immune response. These 3D sSE tissues consistently replicate the molecular pathways found in SSc skin and allow for a controlled model system of SSc to manipulate and test drug therapy responses.
To cite this abstract in AMA style:
Toledo DM, Huang M, Wang Y, Mehta BK, Wood TA, Smith A, Nesbeth Y, Ivanovska I, Christensen B, Pioli PA, Garlick J, Whitfield ML. Molecular Analysis of a Skin Equivalent Tissue Culture Model System of Systemic Sclerosis Using RNA Sequencing, Epigenetic Assays, Histology, and Immunoassays [abstract]. Arthritis Rheumatol. 2018; 70 (suppl 9). https://acrabstracts.org/abstract/molecular-analysis-of-a-skin-equivalent-tissue-culture-model-system-of-systemic-sclerosis-using-rna-sequencing-epigenetic-assays-histology-and-immunoassays/. Accessed .« Back to 2018 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/molecular-analysis-of-a-skin-equivalent-tissue-culture-model-system-of-systemic-sclerosis-using-rna-sequencing-epigenetic-assays-histology-and-immunoassays/