Session Information
Date: Tuesday, November 7, 2017
Title: Plenary Session III
Session Type: ACR Plenary Session
Session Time: 11:00AM-12:30PM
Background/Purpose: Classification and treatment decisions in lupus nephritis (LN) are largely based on renal histology. Single-cell RNAseq (scRNAseq) analysis may accurately differentiate types of renal involvement at the transcriptomic level, and better inform treatment decisions and prognosis. Through our involvement with the accelerating medicines partnership (AMP) network our objectives were to use scRNAseq profiles of renal constituent cells to distinguish between proliferative and membranous LN subclasses, and explore the use of scRNAseq of skin cells to discover biomarkers in a readily obtainable tissue.
Methods: scRNAseq was performed on ~2 mg kidney tissue collected from clinically indicated renal biopsies, and skin biopsies obtained at the time of renal biopsies, in 20 SLE patients. scRNAseq was performed using Fluidigm C1 HT Integrated Fluidic Circuits and cDNA libraries were prepared using the Nextera XT DNA Library Prep Kit followed by NextSeq (Illumina) sequencing.
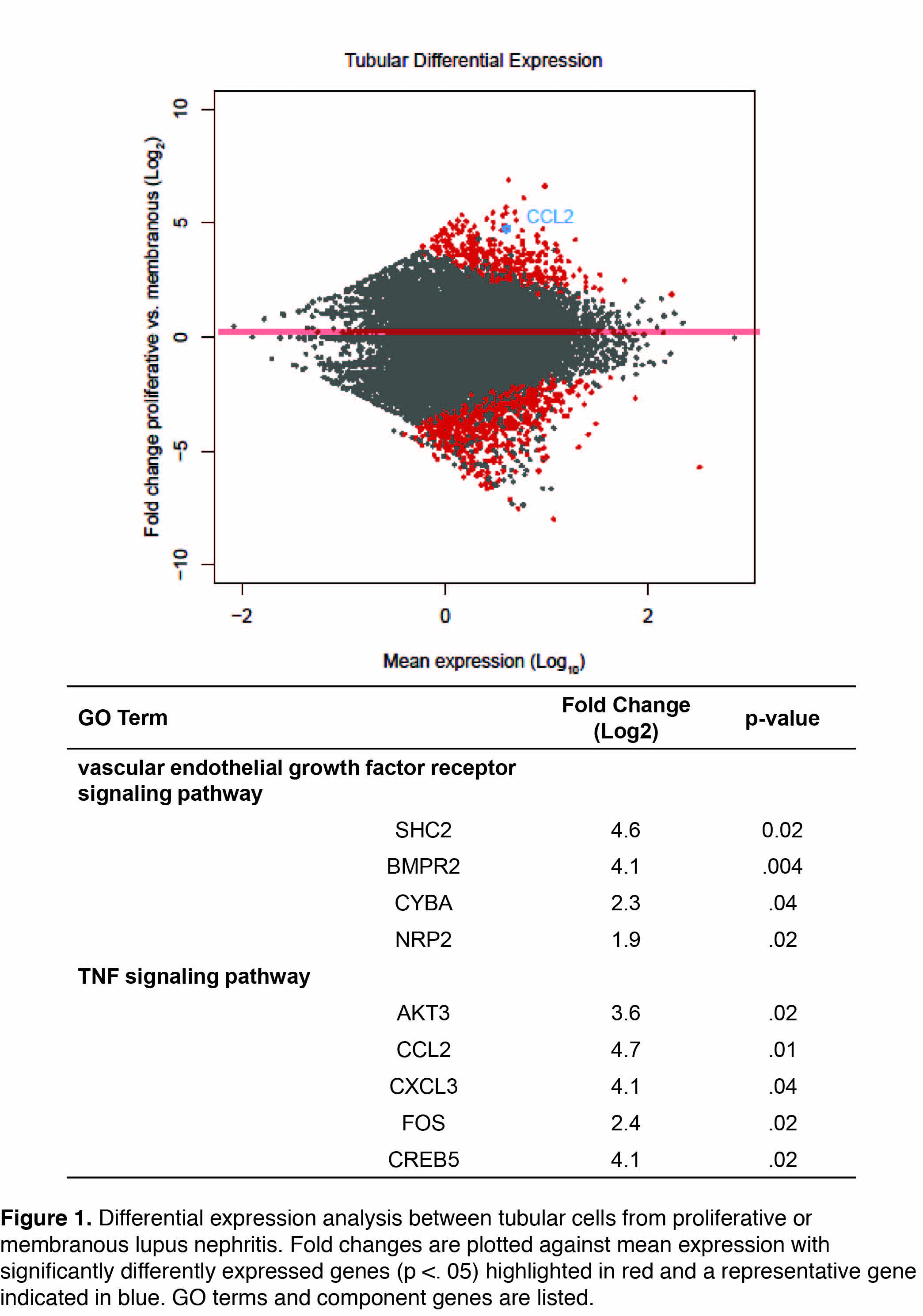
Results: A total of 1616 renal cells and 2392 skin cells were sequenced from LN and healthy control skin and kidney biopsies. Cell-types were determined using principal component analysis and tSNE plotting, resulting in the definitive identification of keratinocytes (N = 2004 cells), tubular cells (N=936 cells), fibroblasts (N=422), endothelial cells (N=154), and leukocytes (N=129). Genes identified by differential expression analysis of tubular cells (Fig. 1) and keratinocytes originating from patients with proliferative (class III or IV) (N=7 patients) and membranous (class V) nephritis (N=6 patients) were subjected to gene ontology pathway analysis. Tubular cells in patients with proliferative nephritis demonstrated upregulated TNF signaling (p<.001), including the transcription factor FOS, as compared to patients with membranous nephropathy. Moreover, the VEGF signaling (p<.001) pathway was upregulated, as was chemokine activity (p<.001) including CCL2 and CXCL3. Interestingly, keratinocytes from non-lesional skin of patients with proliferative nephritis also demonstrated upregulated TNF signaling (p<.01) as compared to those with membranous nephritis.
Conclusion: scRNAseq from small amounts of renal biopsy tissue in SLE can differentiate between the different classes of LN, and provide important insights into potential pathogenic mechanisms. Further, due to the systemic nature of the disease, transcriptomic changes in the skin of LN patients can provide a useful source of biomarkers and may reflect important information concerning concurrent kidney pathological events.
To cite this abstract in AMA style:
Der E, Suryawanshi H, Ranabothu S, Goilav B, Belmont HM, Izmirly PM, Bornkamp N, Jordan N, Wang T, Wu M, James JA, Guthridge JM, Raychaudhuri S, Tuschl T, Buyon JP, Putterman C. Kidney and Skin Single-Cell RNA Sequencing in Lupus Nephritis Provides Mechanistic Insights and Novel Potential Biomarkers [abstract]. Arthritis Rheumatol. 2017; 69 (suppl 10). https://acrabstracts.org/abstract/kidney-and-skin-single-cell-rna-sequencing-in-lupus-nephritis-provides-mechanistic-insights-and-novel-potential-biomarkers/. Accessed .« Back to 2017 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/kidney-and-skin-single-cell-rna-sequencing-in-lupus-nephritis-provides-mechanistic-insights-and-novel-potential-biomarkers/