Session Information
Date: Monday, November 6, 2017
Session Type: ACR Poster Session B
Session Time: 9:00AM-11:00AM
Background/Purpose: Systemic sclerosis (SSc) is a complex systemic autoimmune disease caused by complicated interaction between genetic, epigenetic and environmental risk factors. Evidence showed epigenetic modifications, including DNA methylation, play an important part in the regulation of gene expression and the pathogenesis of a large number of autoimmune diseases such as rheumatoid arthritis (RA) and systemic lupus erythematosus (SLE). However, variations of DNA methylation in SSc have not been comprehensively investigated, especially for subtypes of the immune cells separately. To examine the methylation status in CD4+ and CD8+ T cells of SSc, we performed genome-wide DNA methylation microarray study in Chinese SSc patients and matched controls.
Methods: CD4+ and CD8+ T cells were obtained from SSc patients (n =24) and from age- and sex-matched healthy controls (n =24). All patients met the 2013 criteria for SSc established by the ACR/EULAR. Illumina Infinium HumanMethylation450 BeadChip was utilized for DNA quantification. The significantly differentially methylated sites (DMS) were validated using targeted bisulfite sequencing in an extended cohort consisting of 43 SSc patients and 41 controls (including 12 cases and 12 controls from the initial group). The expression profiles of the differentially methylated genes were measured by RT-PCR. In addition, the serum level of type I interferon-alpha/beta in SSc patients and controls was also quantified by ELISA.
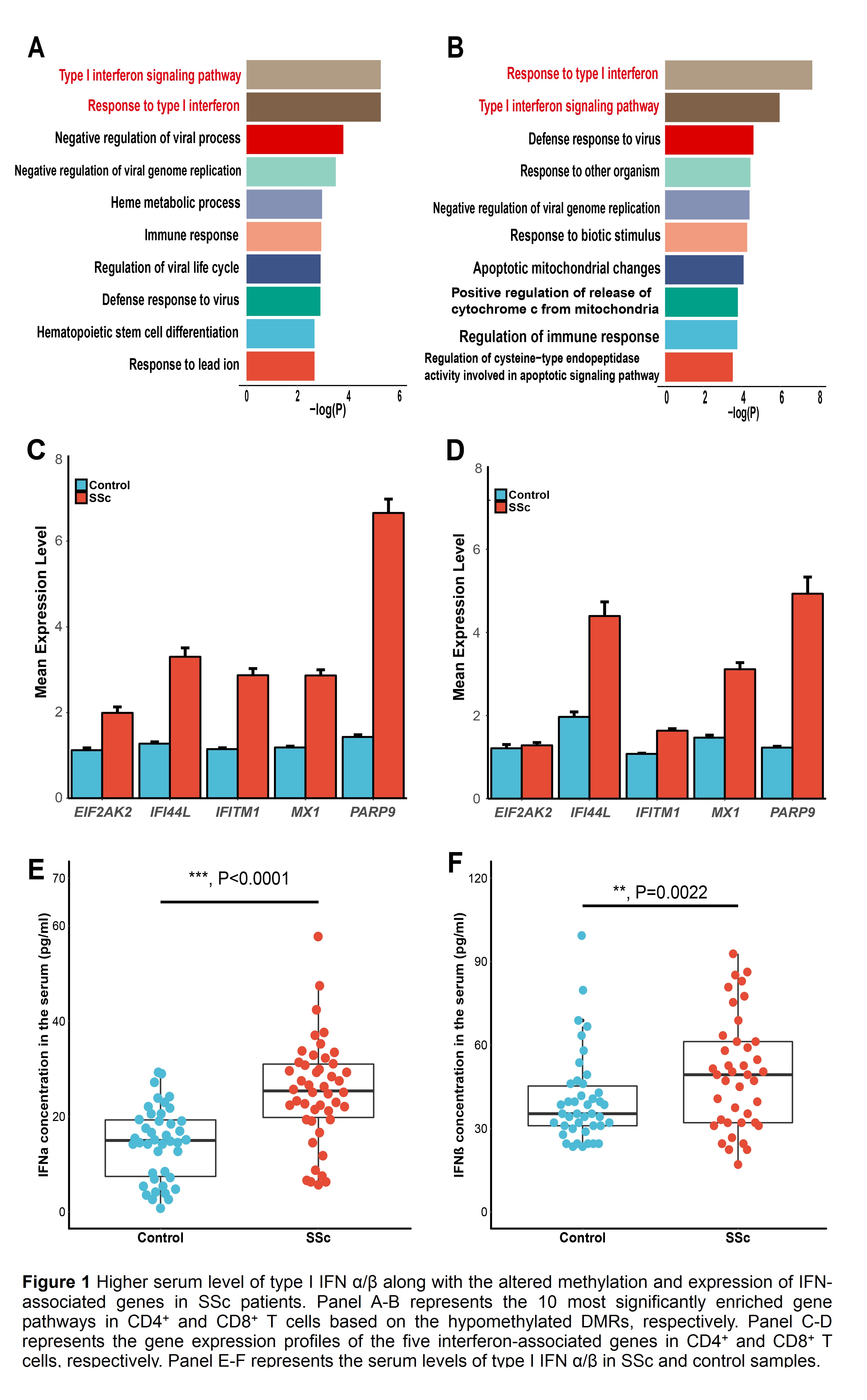
Results: In the discovery stage, type I interferon (IFN)-associated genes were significantly hypomethylated and type I IFN signaling pathway was most significantly enriched in both CD4+ and CD8+ T cells of SSc patients compared to controls, suggesting an abnormality of the type I IFN pathway in SSc patients (Fig 1-A, B). In the second stage, the significantly hypomethylation status of five type I IFN-associated genes (EIF2AK2, IFI44L, IFITM1, MX1, PARP9) was validated (by targeted bisulfite sequencing). Moreover, the upregulation of these genes was also shown in both CD4+ CD8+ T cells of SSc patients (by RT-PCR) (Fig 1-C, D). Further, the serum levels of type I IFN-alpha/beta were also elevated in SSc patients over controls (by ELISA) (Fig 1-E, F). The significant correlations between gene expression, DNA methylation and serum level of type I IFN-alpha/beta were also confirmed, though showing different patterns between CD4+ and CD8+ T cells.
Conclusion: Hypomethylation and upregulation of type I IFN-associated genes were observed in both CD4+ and CD8+ T cells of SSc patients. The serum level of type I IFN-alpha/beta was elevated in SSc patients and correlated significantly with the methylation and expression of its associated genes. It is suggested that hypomethylation of type I IFN-associated genes may be involved in the pathogenesis of SSc.
To cite this abstract in AMA style:
Pu W, Ding W, Wang L, Jiang S, Tu W, Guo S, Liu Q, Ma Y, Chen S, Wu W, Zhou X, Mayes MD, Assassi S, Reveille JD, Jin L, Wang J. Genome-Wide DNA Methylation Analysis in Systemic Sclerosis Reveals Hypomethylation of Interferon-Associated Genes in CD4+ and CD8+ T Cells [abstract]. Arthritis Rheumatol. 2017; 69 (suppl 10). https://acrabstracts.org/abstract/genome-wide-dna-methylation-analysis-in-systemic-sclerosis-reveals-hypomethylation-of-interferon-associated-genes-in-cd4-and-cd8-t-cells/. Accessed .« Back to 2017 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/genome-wide-dna-methylation-analysis-in-systemic-sclerosis-reveals-hypomethylation-of-interferon-associated-genes-in-cd4-and-cd8-t-cells/