Session Information
Session Type: ACR Poster Session B
Session Time: 9:00AM-11:00AM
Background/Purpose: Measurements of immune cell proportions from whole blood can be used to detect pharmacodynamic effects, as a marker for prognosis and to aid in understanding therapeutic response in patients with disease. Profiling of immune cell types from whole blood in clinical trials is traditionally accomplished with low resolution tests such as complete blood count (CBC). Although FACS yields more comprehensive profiling it is challenging to execute in large trials. Mathematical deconvolution techniques have been developed to estimate the proportion of constituent cell types from whole blood using microarray expression profiling (Abbas et al., 2005, Newman et al., 2015) and RNA profiling by next-generation sequencing (NGS) (Gong et al., 2011). Here we extend the technique using RNA-Seq of naïve and activated immune cells to derive cell and activation specific gene signatures.
Methods: Immune cells from lymphoid and myeloid lineages were isolated from multiple healthy donors. Naïve cells were stimulated to induce an activated state. RNA from 102 samples were harvested and profiled by NGS. The deconvolution algorithm leverages a set of genes and associated expression levels to infer cell abundances. Multiple filters were used to ensure specificity and sensitivity during gene identification and matrix generation (Figure 1).
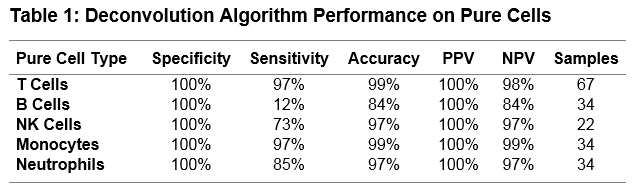
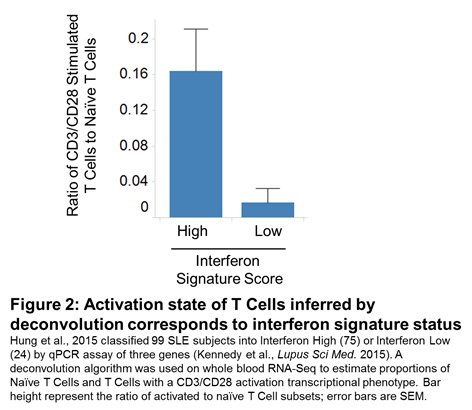
Results: We assessed the ability of the deconvolution algorithm to discriminate pure cell types (T, B, NK, Monocytes and Neutrophils) on an independent RNA-Seq dataset of cells sorted from whole blood from Systemic Lupus Erythematosus (SLE), Multiple Sclerosis and Normal Healthy subjects comprising 191 samples. It achieved an average positive predictive value of 100% and an average negative predictive value of 95% (Table 1). Further validation is planned using whole blood samples analyzed in parallel by FACS and RNA-Seq. We applied this method to a large publically available RNA-Seq dataset of 99 SLE subjects (Hung et al., 2015). We were able to identify a higher proportion of activated T-cells (defined by CD3/CD28 stimulation) in SLE subjects and those corresponded with interferon activity levels as determined by the authors (Figure 2).
Conclusion: Whole blood deconvolution using RNA-Seq may be a useful method for immuno-phenotyping by proxy when FACS is unavailable.
To cite this abstract in AMA style:
Jabado O, Hu S, Carman J, Suchard S, Lee D, Qi Z, Kirov S, Golhar R, He A, Speake C, Linsley PS, Nadler SG, Bandyopadhyay S. Deconvolution of Immune Cell Proportions from Whole Blood RNA Using Next-Generation Sequencing [abstract]. Arthritis Rheumatol. 2016; 68 (suppl 10). https://acrabstracts.org/abstract/deconvolution-of-immune-cell-proportions-from-whole-blood-rna-using-next-generation-sequencing/. Accessed .« Back to 2016 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/deconvolution-of-immune-cell-proportions-from-whole-blood-rna-using-next-generation-sequencing/