Session Information
Date: Sunday, November 12, 2023
Title: Abstracts: B Cell Biology & Targets in Autoimmune & Inflammatory Disease
Session Type: Abstract Session
Session Time: 2:00PM-3:30PM
Background/Purpose: Age-associated B cells (ABCs) accumulate and contribute to the pathogenesis of systemic autoimmune diseases like lupus. Despite recent insights into the ontogeny and function of ABCs, the key transcriptional regulators of ABCs differentiation remain ill-defined. Addressing this gap in knowledge will reveal targets to enable the prevention or reduction of ABCs in disease therapy.
Methods: We employed single-cell RNA sequencing to identify transcription factors (TFs) that were differentially expressed in age-associated B cells (ABCs) from both lupus patients and lupus-prone mice. An arrayed CRISPR screen of these regulatory TFs was conducted under in-vitro conditions that induce ABC formation. In our cohort, we included patients diagnosed with Mowat-Wilson Syndrome (MWS), a condition attributed to ZEB2 haploinsufficiency. We constructed B-cell-specific Zeb2 knockout mice and subjected them to TLR7-driving and Bm12-transfer lupus models, enabling us to study Zeb2’s regulatory role in ABC formation and associated autoimmunity. The detection of autoantibodies was achieved using an autoantigen microarray. To investigate Zeb2’s direct targets, we performed ATAC-seq, Cut & Tag, and Cut & Run sequencing, and integrated these regulome findings with transcriptome data obtained from RNA sequencing. We employed trans-well and PHrodo staining techniques to study the distinct migratory and phagocytic features of ABCs. The induced lupus models were used to evaluate the potential of the JAK1/3 inhibitor tofacitinib in intervening with ABC formation and associated autoimmunity.
Results: We carried out a screening process and identified that Zeb2 is required for in-vitro differentiation of ABCs in both humans and mice. We observed a reduction in ABCs in individuals with ZEB2 haploinsufficiency and in mice devoid of Zeb2 in B-cells. By utilizing mice that selectively lack Zeb2 in B cells, we demonstrated the essential role of Zeb2 in facilitating autoimmune pathology relevant to ABCs, including autoantibody formation, production of proinflammatory cytokines and chemokines, migration to end-organ tissues, and induction of tissue damage in a lupus model. Zeb2 binds to the +20kb intronic enhancer of Mef2b, thereby repressing Mef2b-mediated germinal center B-cell differentiation and instead promoting ABC formation. Zeb2 also targets genes crucial for ABC specification and function, including Itgax. Furthermore, Zeb2 regulates the distinct cellular characteristics of ABCs, including their migration and phagocytic abilities. The differentiation of ABCs driven by Zeb2 necessitates Jak-Stat signaling. Notably, we found that the use of tofacitinib, an approved JAK1/3 inhibitor, effectively reduces the accumulation of ABCs in autoimmune mice and patients.
Conclusion: Our study reveals that ABCs, a unique B cell subset, rely on Zeb2 expression and regulation. Zeb2 affects germinal center B cell development and dictates ABC identity. Modulating Zeb2-mediated Jak-Stat signaling could effectively curb ABC accumulation and related autoimmunity, presenting a potential therapy for autoimmune diseases.
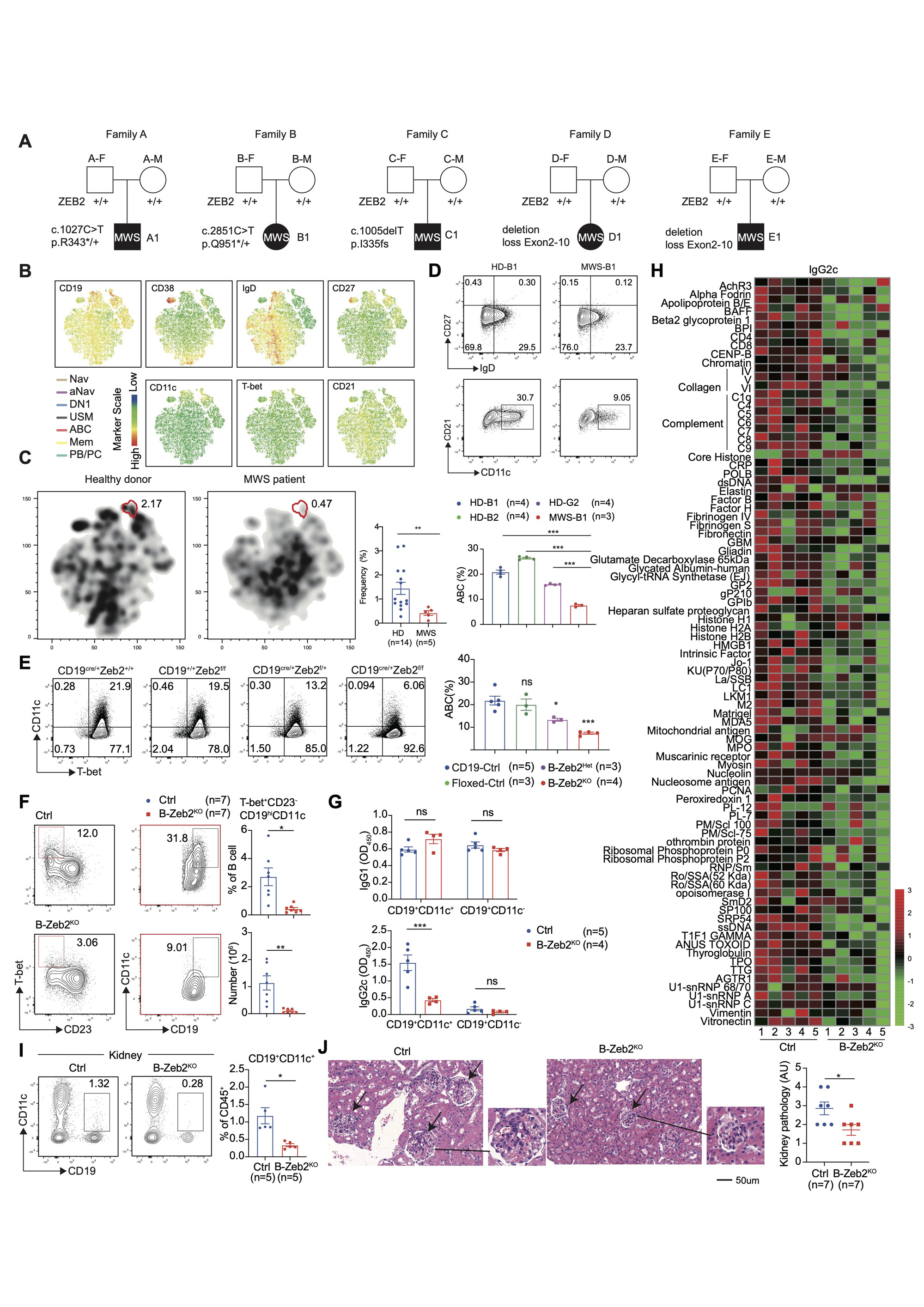
(A) Family pedigrees showing de novo heterozygous mutations of ZEB2 in Mowat-Wilson syndrome (MWS) patients. (B) Comprised t-SNE plots of peripheral B cell clusters for all donors. Eight B cell clusters were identified based on lineage marker expression as naïve B cell (Nav), activate naïve B cell (aNav), CXCR5+ double negative B cells (DN1), unswitch memory B cells (USM), age-associated B cells (ABC), memory B cell (Mem), plasma blast (PB) and plasma cells (PC). (C) Separated t-SNE plots for two representative HD (left) and MWS (right) samples. Statistical analysis of ABC in PBMC for HD versus MWS group. (D) Primary B cells were isolated from HDs and a MWS patient and cultured with an ABC-skewing cocktail for 3 days. Representative plots and frequency of ABCs (CD19+CD38-CD27-IgD-CD11c+CD21-). (E) Splenic B cells were isolated from CD19Cre/+ (CD19-Ctrl), Zeb2f/f (Floxed-Ctrl), Zeb2f/+CD19cre/+(B-Zeb2Het), Zeb2f/f CD19cre/+(B-Zeb2KO) mice and cultured with ABC-skewing cocktail for 3 days. Representative plots and frequency of ABCs (CD19+CD11c+T-bet+). (F) B-Zeb2KO and CD19Cre/+ (Ctrl) mice were induced by imiquimod (IMQ) for 6 weeks. Flow cytometry plots and statistical analysis of splenic ABCs (CD19+CD23-CD11c+T-bet+) (G) Splenic CD19+CD21-CD11c+ and CD19+CD21+CD11c- B cells sorted by flow cytometry from mice described in (F) were stimulated with anti-IgM, anti-CD40, R848, IL_21 for one day and then the supernatants were collected to detect IgG1 and IgG2c antibodies. (H) Autoantigen microarray was used to detect IgG2c autoantibodies in the serum of mice described in (F) by OmicsArray. (I) Representative plots and frequency of renal ABCs (CD19+CD11c+) from mice described in (F). (J) H&E staining (left) analysis of kidneys from mice described in (F). Bars indicate mean ± SEM values. Statistical significance determined by Student’s t-test.*p﹤0.05, **P﹤0.01, ***P﹤0.001, ns, not significant.
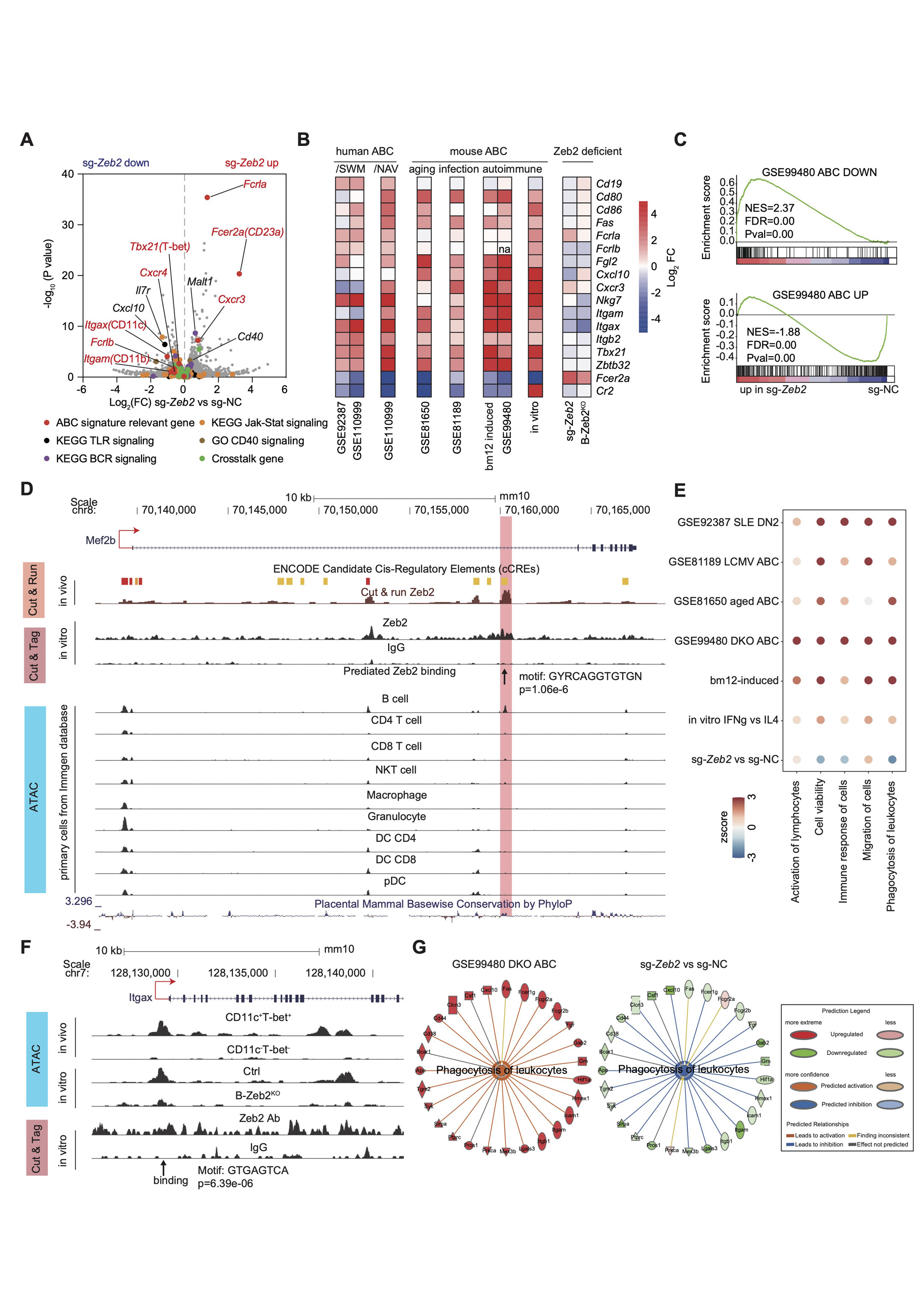
(A) Volcano graph showing the transcriptional profiles in mouse B cells edited by sg-NC and sg-Zeb2 under in vitro ABC induction for 3 days. ABC signature relevant genes, genes of BCR, TLR, Jak-Stat signaling, CD40 signaling and crosstalk genes were labeled with colored dots. (B) Heatmap showing expression of representative ABC signature genes in publicly available RNA-seq datasets: GSE92387, GSE110999, GSE81650, GSE81189, GSE99480 and our own RNA-seq datasets: bm12-induced (Splenic CD11c+T-bet+CD21- and CD11c-T-bet-CD21+ B cells were sorted by flow cytometry from T-bet-tdTOMATO reporter mice immunized with bm12 T cells), in vitro (mouse B cells were cultured with ABC-skewing cocktail for 3 days), sg-Zeb2 (described in (A)), B-Zeb2KO (B cells from B-Zeb2KO and Ctrl mice were cultured with ABC-skewing cocktail for 3 days). (C) GSEA showing the enrichment of the ABC-down geneset and ABC-up geneset from DKO mice (GSE99480) in sg-Zeb2 versus sg-NC ABCs. (D) CUT & RUN, CUT & Tag, and ATAC-seq tracks display Zeb2 binding around the Mef2b locus in indicated immune cells, visualized with the UCSC genome browser. The chromatin accessibility of Mef2b in mouse primary immune cell subsets from Immgene database. (E) Chromatin accessibility by ATAC-seq and Zeb2-binding by CUT & Tag in the promoter site of mouse Itgax. ATAC-seq was performed in ex-vivo sorted ABC (CD11c+T-bet+) and non-ABC (CD11c-T-bet-) from bm12-induced mice and in-vitro derived Zeb2-deficient and ctrl B cell cultured for 3 day under ABC skewing condition. (F) Dot plot showing the activation Z-score for IPA-predicated biological function of ABCs including activation, cell viability, immune response, migration and phagocytosis from RNA-seq datasets described as in (B). (G) Network diagram representing phagocytosis pathway in sg-Zeb2 versus sg-NC ABCs by IPA. The color of each node indicates change in the expression: red (upregulated) and green (downregulated). The edges indicate the predicted relationship between nodes and biological function: orange representing activation, blue representing inhibition, gray representing effect not predicted.
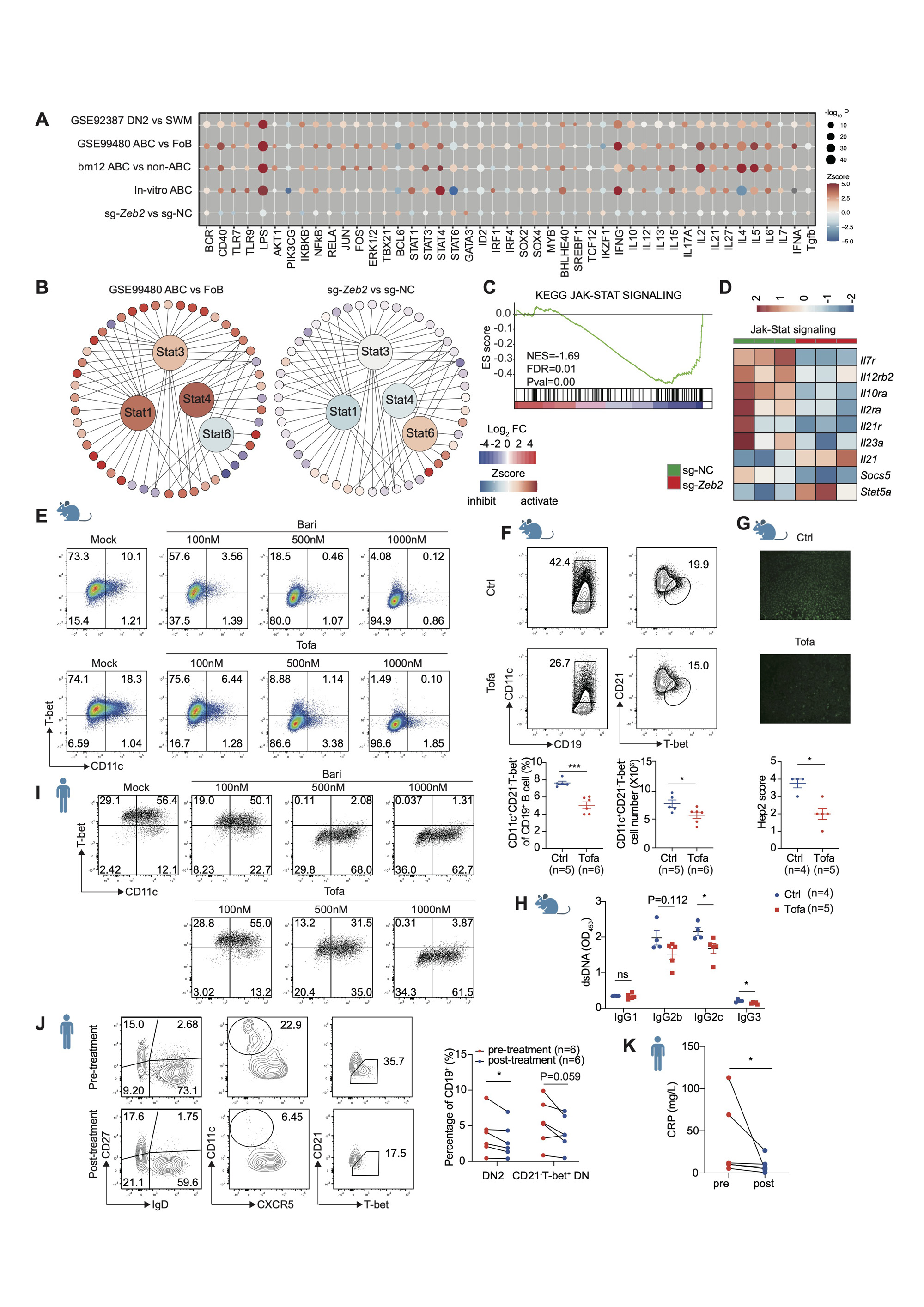
(A) Activation Z-score heatmap for IPA-predicted upstream regulators in indicated datasets mainly described in Fig 2B. P-values were log-normalized and presented by the size of the plot. The color of the plot indicates the activated (orange) or inhibited (blue) regulation of the predicted regulators. (B) Upstream regulator analysis showing Stat1, Stat3, Stat4 activated and Stat6 inhibited in ABCs (DKO, GSE99480), while the opposite effects were observed in sg-Zeb2 vs sg-NC dataset. The color of the surrounding circles indicates the change in the expression: red (upregulated) and blue (downregulated). The color of the center circles indicates predicted regulators: orange (activated) and blue (inhibited). (C) GSEA showing impaired Jak-Stat pathway in sg-Zeb2+ B cells compared with sg-NC. (D) Heatmap showing expression of selected Jak-Stat signaling genes in sg-NC+ and sg-Zeb2+ B cells. (E) Flow cytometry plots of in vitro induced mouse ABCs (CD11c+T-bet+) with different concentrations of baricitinib (Bari) or tofacitinib (Tofa). (F-H) C57 mice were immunized with bm12 T cells for 2 weeks and treated with tofacitinib (Tofa) or dissolved solution (Ctrl) by oral gavage once a day to analyze splenic ABCs (CD19+CD11C+T-bet+CD21-, F), ANA (G) and anti-dsDNA Ig (H) in serum. (I) Flow cytometry plots of in vitro induced human ABCs (CD11c+T-bet+) with different concentration of baricitinib or tofacitinib. (J) Flow cytometry plots of human ABCs (DN2, CD19+CD27-IgD-CD11c+CXCR5-) or (CD21-T-bet+ DN, CD19+CD27-IgD-CD21-T-bet+) in PBMCs from patients with rheumatoid arthritis before and after Jak-Stat inhibitor tofacitinib treatment for 4 weeks. (K) CRP level in RA patients described in (J). n represents distinct samples (biological repeats). Data are representative of 2_3 independent experiments. Data are mean ± SEM values. *p<0.05, **P<0.01, ***P<0.001, ns, not significant, unpaired t-test (F, H), Mann-Whitney test (G), paired t-test (J) and paired Wilcoxon test (K).
To cite this abstract in AMA style:
Dai D, Gu S, Han X, Ding H, Jiang Y, Chen S, Shen N. ZEB2 Acts as a Crucial Transcriptional Regulator Governing Age-Associated B Cell Formation and Pathogenicity in Systemic Lupus Erythematosus [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/zeb2-acts-as-a-crucial-transcriptional-regulator-governing-age-associated-b-cell-formation-and-pathogenicity-in-systemic-lupus-erythematosus/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/zeb2-acts-as-a-crucial-transcriptional-regulator-governing-age-associated-b-cell-formation-and-pathogenicity-in-systemic-lupus-erythematosus/