Session Information
Session Type: Poster Session B
Session Time: 10:30AM-12:30PM
Background/Purpose: Eosinophilic granulomatosis with polyangiitis (EGPA) is a vasculitis that can present indolently. This can lead to a delay in diagnosis and treatment. This study used machine learning to identify pre-diagnostic attributes of EGPA, estimate the prevalence of EGPA, and create a clinical decision screening tool to reduce time to diagnose EGPA.
Methods: Leveraging IQVIA’s United States medical and pharmacy claims data and laboratory results, ~2,800–4,000 patients were split into positive and negative cohorts. All patients were required to have a history of asthma or oral glucocorticoid use. Two cohort designs and decision tree models were developed to create a clinical decision screening tool: i) A rolling cross-section (RCS) model that used 2 years of training data and a gap of 0–6 months between end of source data and date of diagnosis in known medical history; ii) A non-RCS model that used ≥1 year of training data and the maximum possible data after January 2016, including claims data available directly before diagnosis.
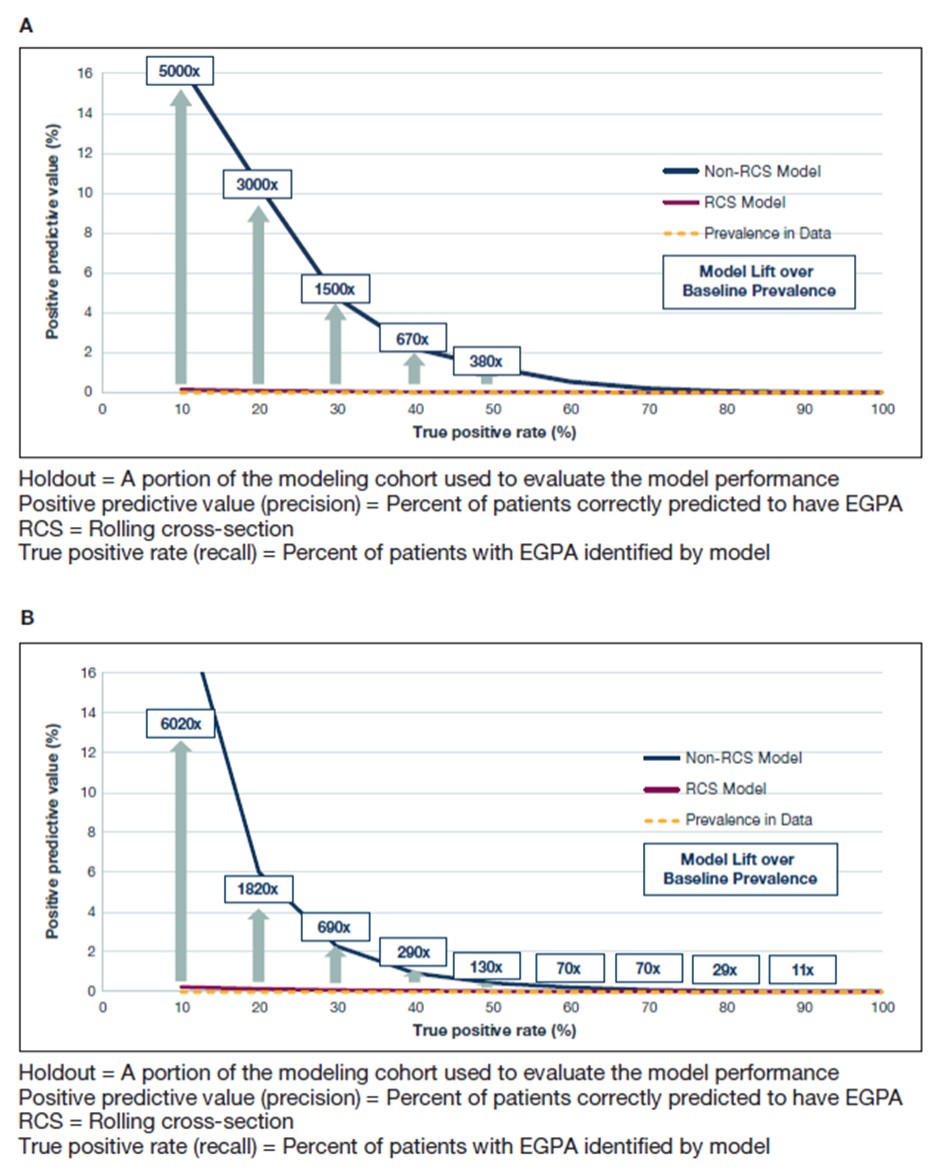
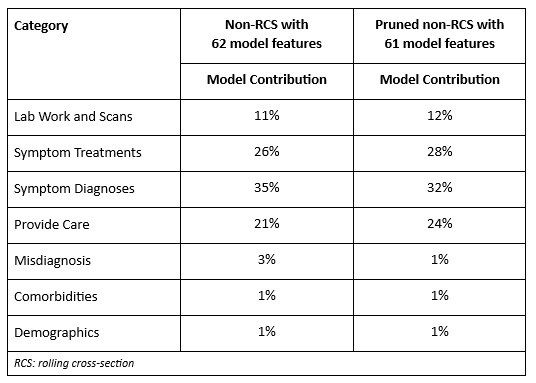
Results: The positive predictive value of all models exceeded the baseline prevalence and incidence rates in the dataset (Figure 1). The RCS model estimated approximately 9,644–11,793 cases of EGPA. Key determinants within the models showed that eosinophilia, asthma, chronic sinusitis, multiple chest x-rays, skin biopsies, and multiple prescriptions of oral glucocorticoids were most predictive of a diagnosis of EGPA (Table 1). The first decision tree model uncovered four possible decision pathways for use within the clinical decision screening tool.
Conclusion: These results demonstrate that leveraging machine learning and claims data can improve the estimation of prevalence of EGPA in the United States and aid in arriving at an earlier diagnosis of EGPA. Up to 100 key events were identified by the model as pivotal in identifying patients with EGPA. The decision tree models may offer improved methods for research using electronic health records and claims data, and help health systems improve the care of patients with EGPA.
To cite this abstract in AMA style:
Merkel P, Baudy P, Carstens D, North B, Danka M, Roy S, Marshall H, Chupp G. Utilizing Machine Learning with Claims Data to Diagnose and Quantify the Prevalence of Eosinophilic Granulomatosis with Polyangiitis [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/utilizing-machine-learning-with-claims-data-to-diagnose-and-quantify-the-prevalence-of-eosinophilic-granulomatosis-with-polyangiitis/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/utilizing-machine-learning-with-claims-data-to-diagnose-and-quantify-the-prevalence-of-eosinophilic-granulomatosis-with-polyangiitis/