Session Information
Session Type: Poster Session A
Session Time: 10:30AM-12:30PM
Background/Purpose: Antiphospholipid syndrome (APS) is a rare autoimmune disease that is likely to demonstrate improved outcomes with earlier diagnosis and treatment. However, given APS’s complex, multi-system disease manifestations, as well as the likelihood of underrecognition by some community-based providers, patients may not have received the proper testing and/or referrals to receive a diagnosis. Here, we present a first-generation prediction model for APS which does not rely on APS-specific diagnostic codes or laboratory tests, and can potentially be used to identify patients with undiagnosed APS.
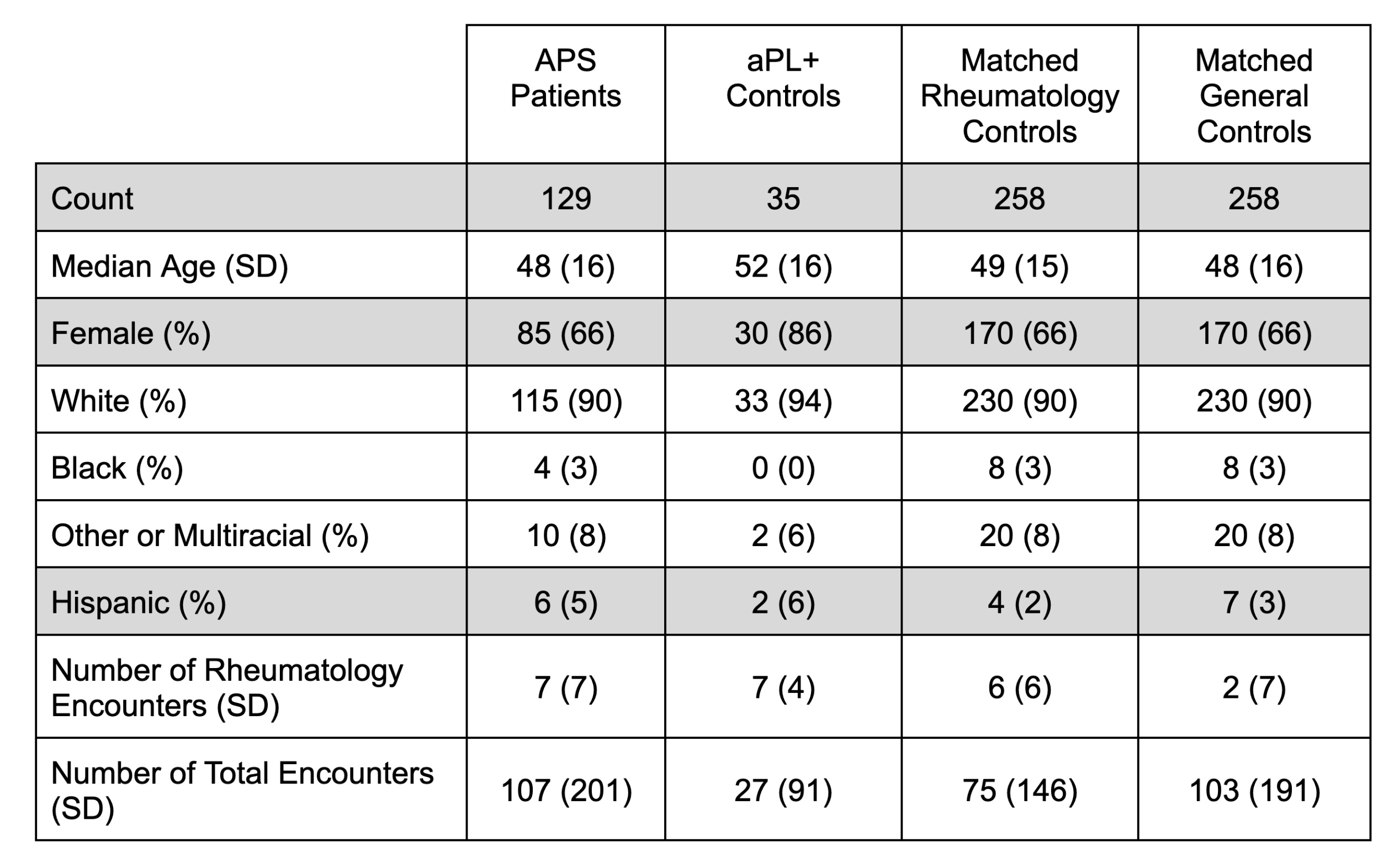
Methods: Electronic health record (EHR) data (2015-2023) from a single United States-based academic medical center were used. The study population included 129 APS patients, whose classification was verified by APS experts. The control groups included 35 antiphospholipid antibody (aPL)-only patients (who had positive aPL tests, but did not meet the classification criteria for APS) and 258 individuals matched for demographics and healthcare utilization (Table 1). Structured EHR data for ICD-10 codes, medications, and laboratory tests were engineered into 1,878 features with guidance from APS experts. APS-specific features that were excluded from the model included certain ICD-10 codes (D68.61, D68.312, D68.62) and laboratory tests (anticardiolipin, anti-beta-2 glycoprotein I, and lupus anticoagulant). A gradient-boosted decision tree (GBDT) from the xgboost R package (version 1.7.7.1) with max_depth = 14 was trained to classify APS vs. all controls and evaluated on a held-out test set.
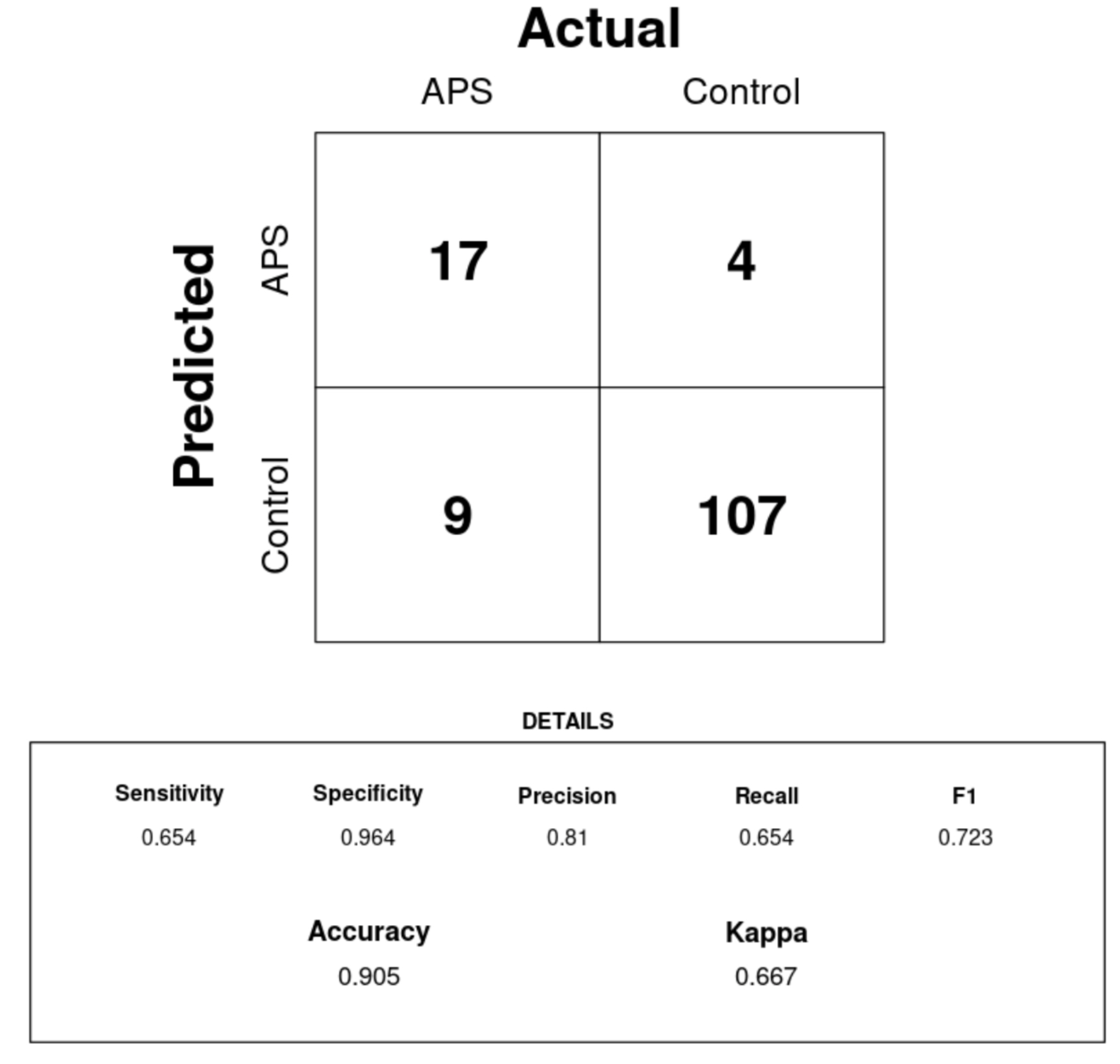
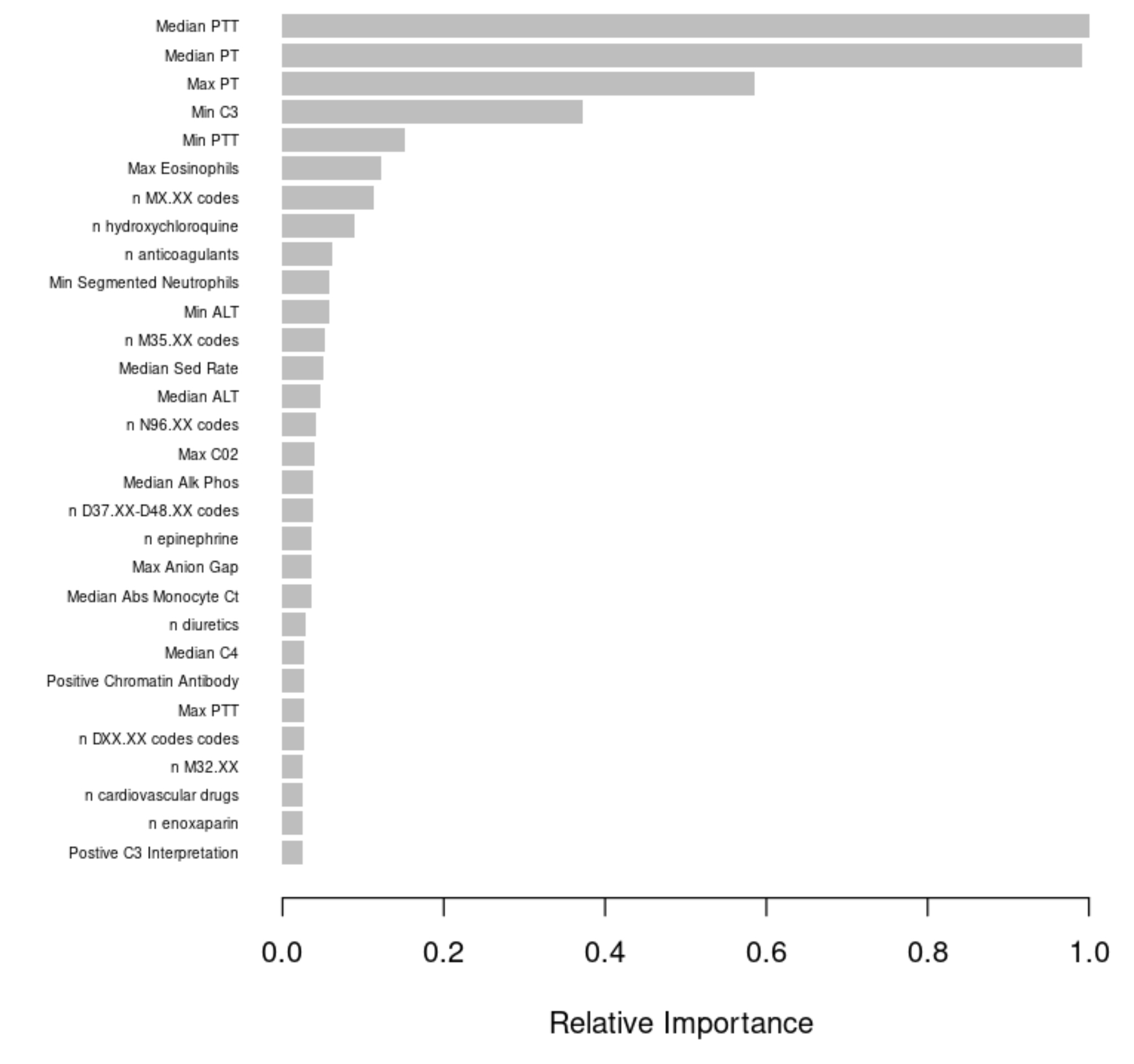
Results: Our GBDT model to classify patients as having or not having APS without APS-specific features had strong performance on our test set (Figure 1). While the model had only moderate sensitivity (0.65), it had a high specificity (0.96) and precision (0.81). Therefore, the model was able to leverage an APS-specific signature of EHR variables to appropriately classify many APS patients in our sample. While some of this strong performance may have been driven by the high feature importance (Figure 2) of anticoagulant use (which could be the result of an APS diagnosis, not a precursor to it), we believe this first-generation model may still be applicable to diagnosis-naive patients because it captures a breadth of APS-related features like the ICD-10 N96 code for recurrent pregnancy loss and various laboratory tests not influenced by anticoagulants such as segmented neutrophil count.
Conclusion: Earlier APS diagnosis is a promising avenue for improving patient outcomes. This abstract presents proof of concept that APS patients have a signature of EHR data that can be used to identify APS patients who either lack a diagnostic code or a diagnosis altogether. Work is now underway to test the utility of adding more features to the model (e.g., unstructured clinical notes) and, importantly, to perform internal validation of possible cases in our health system via chart review and external validation at other healthcare centers. This model could inform future clinical-decision support tools which suggest to providers (e.g., primary care doctors) that they should order specific blood tests or refer a patient for expert evaluation of APS.
To cite this abstract in AMA style:
Balczewski E, Ambati A, Liang W, Madison J, Zuo Y, Singh K, Knight J. Underdiagnosis Prediction Fingerprint for Antiphospholipid Syndrome Derived from Electronic Health Record Data [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/underdiagnosis-prediction-fingerprint-for-antiphospholipid-syndrome-derived-from-electronic-health-record-data/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/underdiagnosis-prediction-fingerprint-for-antiphospholipid-syndrome-derived-from-electronic-health-record-data/