Session Information
Session Type: Abstract Session
Session Time: 3:00PM-4:30PM
Background/Purpose: Sjögren’s disease (SjD) is a clinically and biologically heterogeneous disease. To date, no phase-III trial showed efficacy in reducing the symptoms or systemic activity of the disease and no immunomodulatory drug has been marketed. We have previously demonstrated using whole blood RNAseq data from the PRECISESADS project that SjD patients can be clustered into 4 distinct groups (endotypes).1 Cluster 1 was characterized by a unique interferon (IFN) signature, cluster 2 comprised healthy-like patients, cluster 3 was marked by IFN and B-cell signatures, cluster 4 was characterized by IFN, B-cell and neutrophil signatures. In this study, we hypothesized that patients in different clusters may have different responses to different drugs due to different therapeutic targets in each cluster
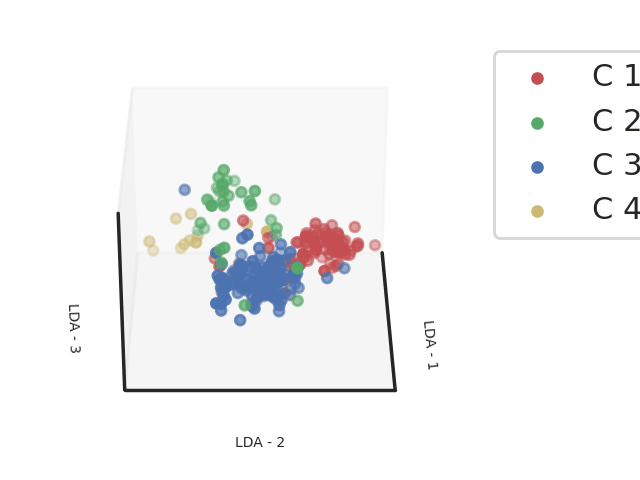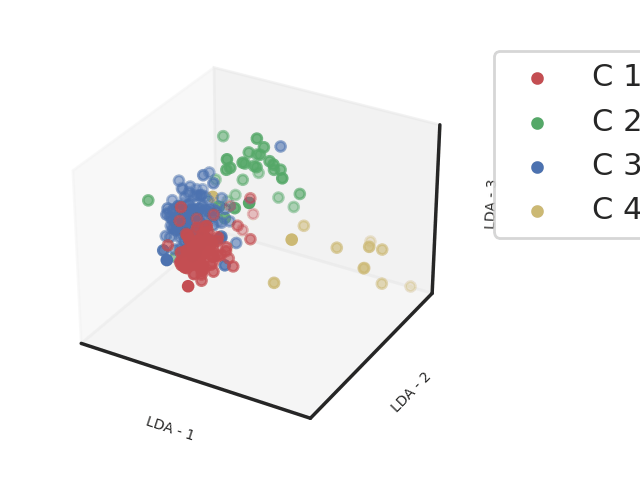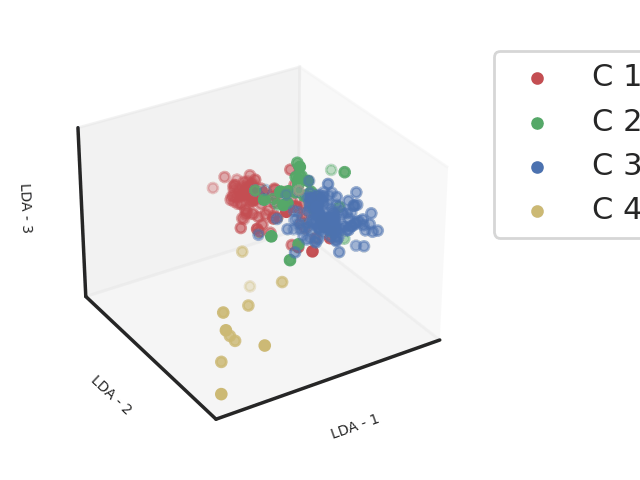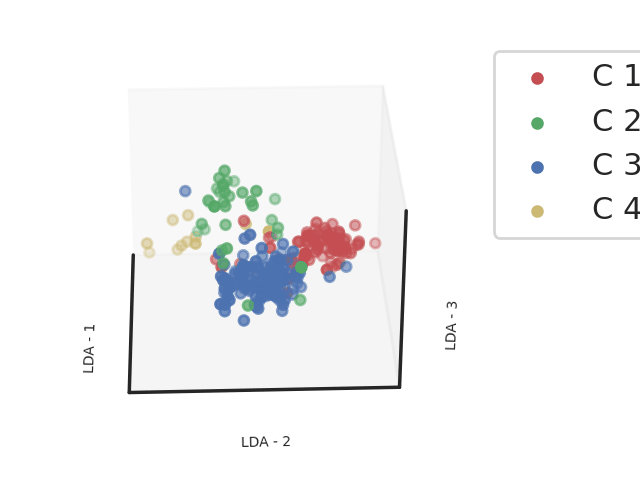
Methods: Clinical, biological and transcriptomic data were obtained from three randomized controlled trials assessing hydroxychloroquine and leflunomide2 (HCQ-LEF, including in our study 13 patients on HCQ-LEF and 5 on placebo), rituximab3 (RTX, of whom 31 patients on RTX and 25 with placebo were included) or abatacept4(ABA, including 58 patients on ABA and 59 on placebo) versus placebo. Whole blood RNAseq was performed on samples collected at inclusion in the clinical trials. Patients were clustered into 4 endotypes using a semi-supervised Uniform Manifold Approximation and Projection for Dimension Reduction (UMAP) algorithm combined with a support vector machine (SVM) trained in respect to our previous work (Figure 1). We compared demographic features and disease characteristics between the 4 clusters. We then assessed response to therapy, defined by the STAR responder index5, in the active treatment and placebo arms within each cluster. We analyzed this response rate in the 3 pooled trials, and then in each clinical trial individually.
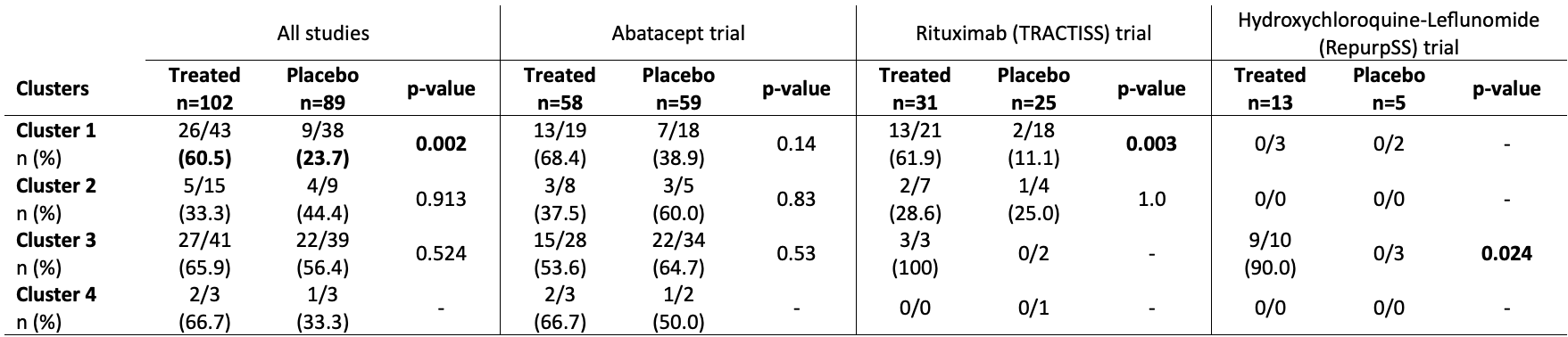
Results: Eighty-one subjects were clustered into cluster 1 (5, 39 and 37 from the HCQ-LEF, RTX and ABA trials, respectively), 24 were in cluster 2 (11 and 13 from the RTX and ABA trials), 80 were in cluster 3 (13, 5 and 62 from the HCQ-LEF, RTX and ABA trials) and 6 patients were in cluster 4 (1 and 5 from the RTX and ABA trials). ESSPRI scores did not differ between the 4 clusters (5.71 to 7.22; p=0.48). Unlike, ESSDAI was lowest in clusters 1 and 2 (6.36 and 6.62, respectively) and significantly higher in clusters 3 and 4 (8.81 and 10.67; p=0.003).
When all 102 treated patients were pooled and compared to the 89 placebo patients, treated patients in cluster 1 were more likely to achieve a STAR response score than control patients (60.5% vs. 23.7%, p=0.002) (Table 1). A similar trend was seen in the RTX trial. In the HCQ-LEF trial, patients within cluster 3 who received the treatment were more likely to reach the STAR score. Whether the studies were pooled or analyzed separately, no treatment compared to placebo showed significant efficacy within cluster 2 (transcriptionally healthy-like patients).
Conclusion: The analysis of controlled trials with transcriptomic-based clusters of SjD patients highlights the lack of response to HCQ-LEF, RTX and ABA in the healthy-like patients. We suggest that these patients are less likely to respond according to STAR score, they may not be included into further controlled trials.
Abbreviations: LDA: Linear Discriminant Analysis
Footnotes: Linear Discriminant analysis of transcriptomic profiles of the patients from 3 different cohorts based on the clustering obtained from a support vector machine classifier
To cite this abstract in AMA style:
Chevet B, Devauchelle V, Pontarini E, Baloche V, Bombardieri M, Bowman S, Barnes M, Sreih A, Liu J, Kelly S, Christodoulou A, Badani H, Moigeon P, LAIGLE L, Soret P, Le dantec C, Pers J, Alarcon-Riquelme M, Barturen G, Mariette X, Van Roon J, Seror R, Nocturne G, Cornec D, Foulquier N. Transcriptomic Stratification Predicts Response to Rituximab, Abatacept or the Association of Hydroxychloroquine and Leflunomide in 3 Randomized Controlled Clinical Trials in Sjögren’s Disease [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/transcriptomic-stratification-predicts-response-to-rituximab-abatacept-or-the-association-of-hydroxychloroquine-and-leflunomide-in-3-randomized-controlled-clinical-trials-in-sjogrens-disease/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/transcriptomic-stratification-predicts-response-to-rituximab-abatacept-or-the-association-of-hydroxychloroquine-and-leflunomide-in-3-randomized-controlled-clinical-trials-in-sjogrens-disease/