Session Information
Session Type: Abstract Session
Session Time: 2:00PM-3:30PM
Background/Purpose: Lupus Nephritis (LN) significantly reduces the survival and life expectancy of patients with SLE. Given this, considerable effort has gone into characterizing the histologic classes that confer the highest standardized mortality ratios. Although Class II LN is considered a milder form of disease which often requires less aggressive treatment, a growing body of clinical evidence suggests that these patients can progress to more advanced LN classes. Accordingly, this study leveraged the multi-center LN Accelerating Medicine Partnership (AMP) Network to focus on the transcriptome of Class II kidney biopsies with comparison to other histologic classes to provide insights into the molecular underpinnings of early disease and potential drivers of progression.
Methods: LN patients were enrolled in AMP at the time of a clinically indicated kidney biopsy and followed for one year. scRNA-seq was performed using the 10x genomics sequencing platform and quality control was performed to retain high-quality cells. Using the AMP consortium dataset which includes patients with membranous, mixed and proliferative LN, a reference-based mapping approach was employed to identify shared cell states and insights into Class II pathogenesis and outcomes.
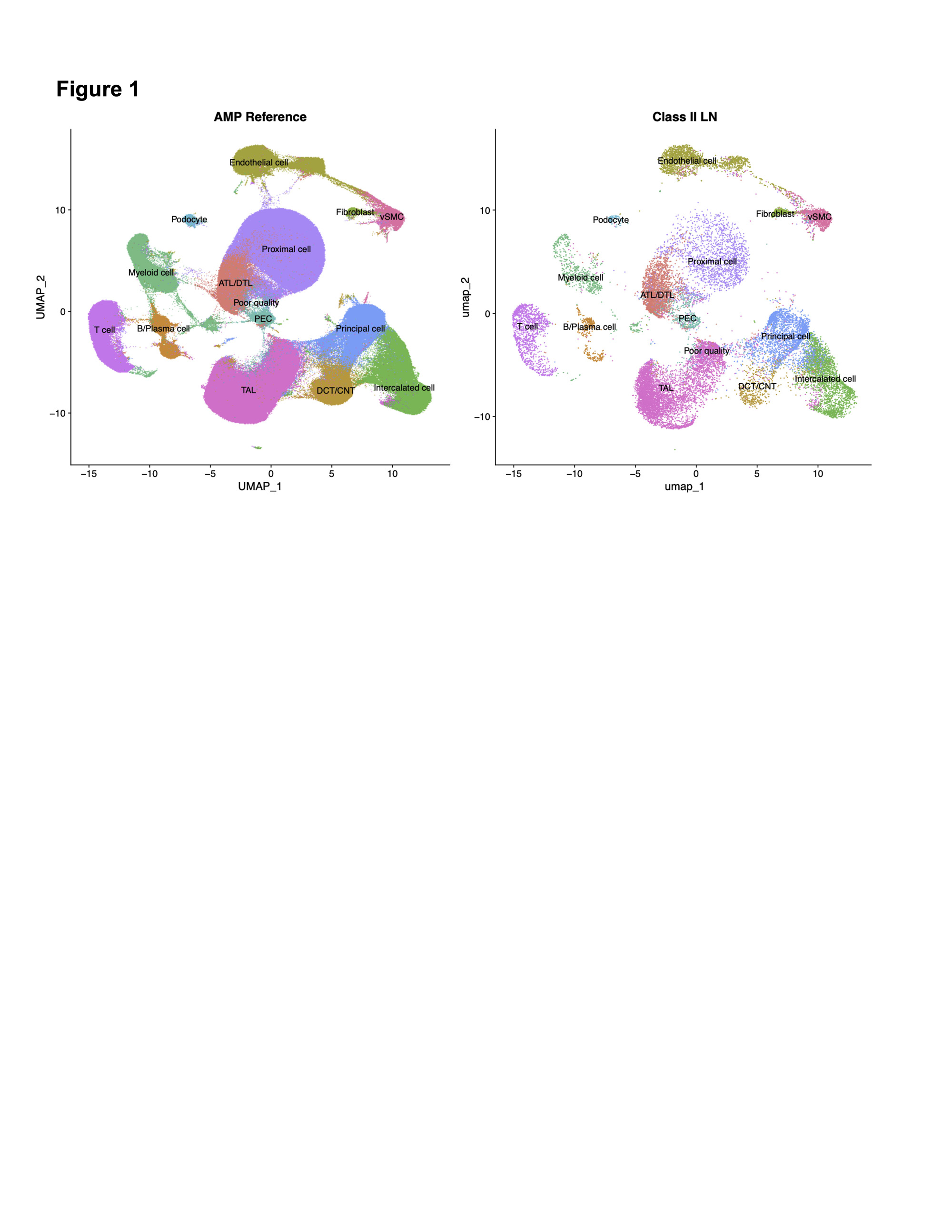
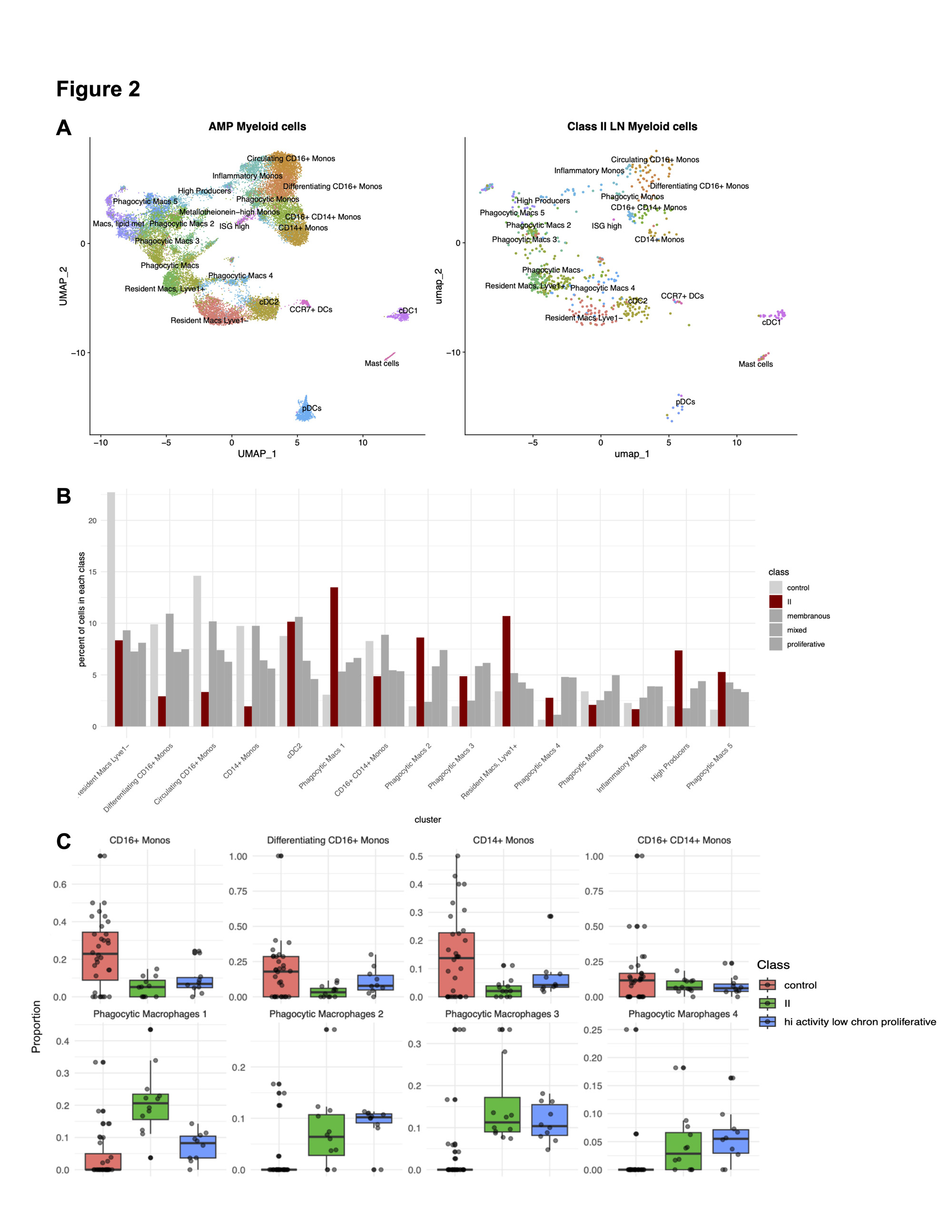
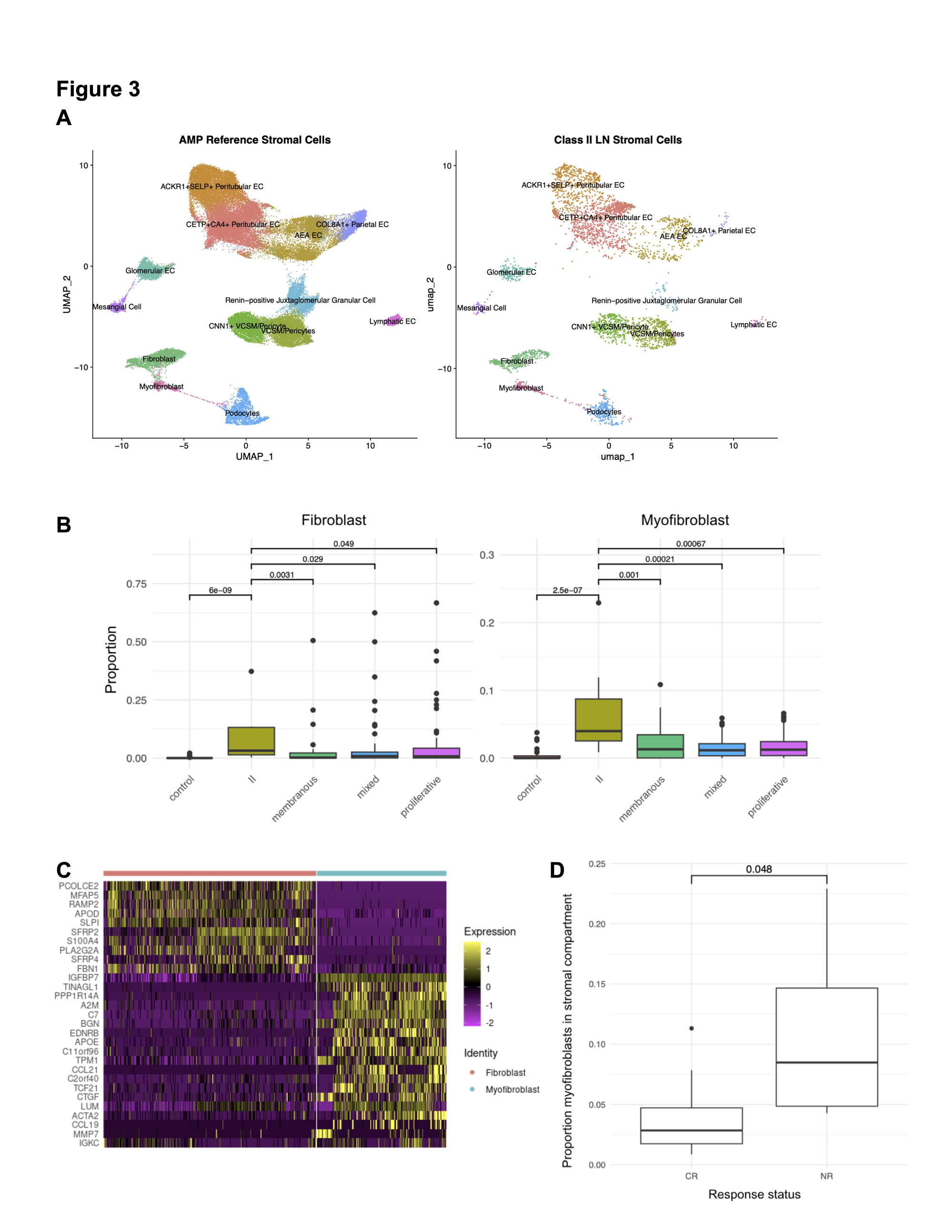
Results: scRNA-Seq from 12 patients with Class II LN biopsies yielded 25,483 high-quality cells comprised of 11 parenchymal and three immune cell types (Fig 1). Reference mapping identified B, T and myeloid cells which are infrequently found in Class II LN by conventional microscopy (Fig 2A). In the myeloid compartment, cell type proportion analysis revealed a distinct signature with fewer monocytes and a greater number of differentiated and phagocytic macrophages in Class II compared to control and other LN classes (Fig 2B). This distinct myeloid signature was also identified in proliferative LN samples with high activity and low chronicity (activity index >= 4, chronicity index = 0). In both Class II and proliferative LN with high activity, monocytes and phagocytic macrophage subtypes were contracted and expanded relative to biopsies from healthy donors, respectively (Fig. 2C). The stromal compartment of Class II unexpectedly had a higher proportion of fibroblasts and myofibroblasts compared to all other classes (3A-B). These myofibroblasts were characterized by high expression of canonical markers, such as ACTA2 as well as markers of immune activation such as CTGF, CCL19, and CCL21 (3C). The proportion of myofibroblasts was higher in Class II patients whose UPCR fell below 0.5 and creatinine remained normal or did not exceed 125% of baseline compared to Class II patients considered non-responders at 52 weeks (3D, p = 0.048).
Conclusion: These data support that histology provides an incomplete picture of the molecular and cellular landscape in Class II LN. Increased phagocytic macrophage activity and immune-induced myofibroblast differentiation were associated with higher disease activity, highlighting these cellular composition changes as potential drivers of non-responsiveness and progression.
To cite this abstract in AMA style:
Shwetar J, Preisinger K, Zaminski D, Carlucci P, Deonaraine K, Xiao Q, Mears J, Gurajala S, peter I, James J, Guthridge J, Fava A, Rovin B, DeJager W, Wu M, Rao D, Putterman C, Diamond B, Fine D, Monroy-Trujillo J, Haag K, Belmont H, Apruzzese W, Davidson A, Payan-Schober F, Furie R, Hoover P, Berthier C, Dall'Era M, Cho K, Kamen D, Kalunian K, Anolik J, Raychaudhuri S, Hacohen N, Petri M, Clancy R, Wofsy D, Arazi A, Ruggles K, Buyon J, SLE/RA T. Transcriptomic Characterization of Class II Lupus Nephritis and Outcomes [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/transcriptomic-characterization-of-class-ii-lupus-nephritis-and-outcomes/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/transcriptomic-characterization-of-class-ii-lupus-nephritis-and-outcomes/