Session Information
Session Type: Poster Session B
Session Time: 9:00AM-10:30AM
Background/Purpose: Predicting the course or response to treatment of lupus nephritis (LN) from standard renal biopsies is problematic. We, therefore sought to understand the molecular endotypes of LN by analyzing bulk RNA from renal biopsies.
Methods: Gene expression of biopsies from 76 kidneys derived from patients with LN was analyzed for enrichment of informative modules of co-expressed genes using Gene Set Variation Analysis (GSVA) and grouped into four clusters with hierarchical clustering. Gene modules identifying immune/inflammatory cells as well as resident kidney cells and metabolic activities were employed. Kidney biopsy histology and immunofluorescence were scored by a blinded clinical pathologist.
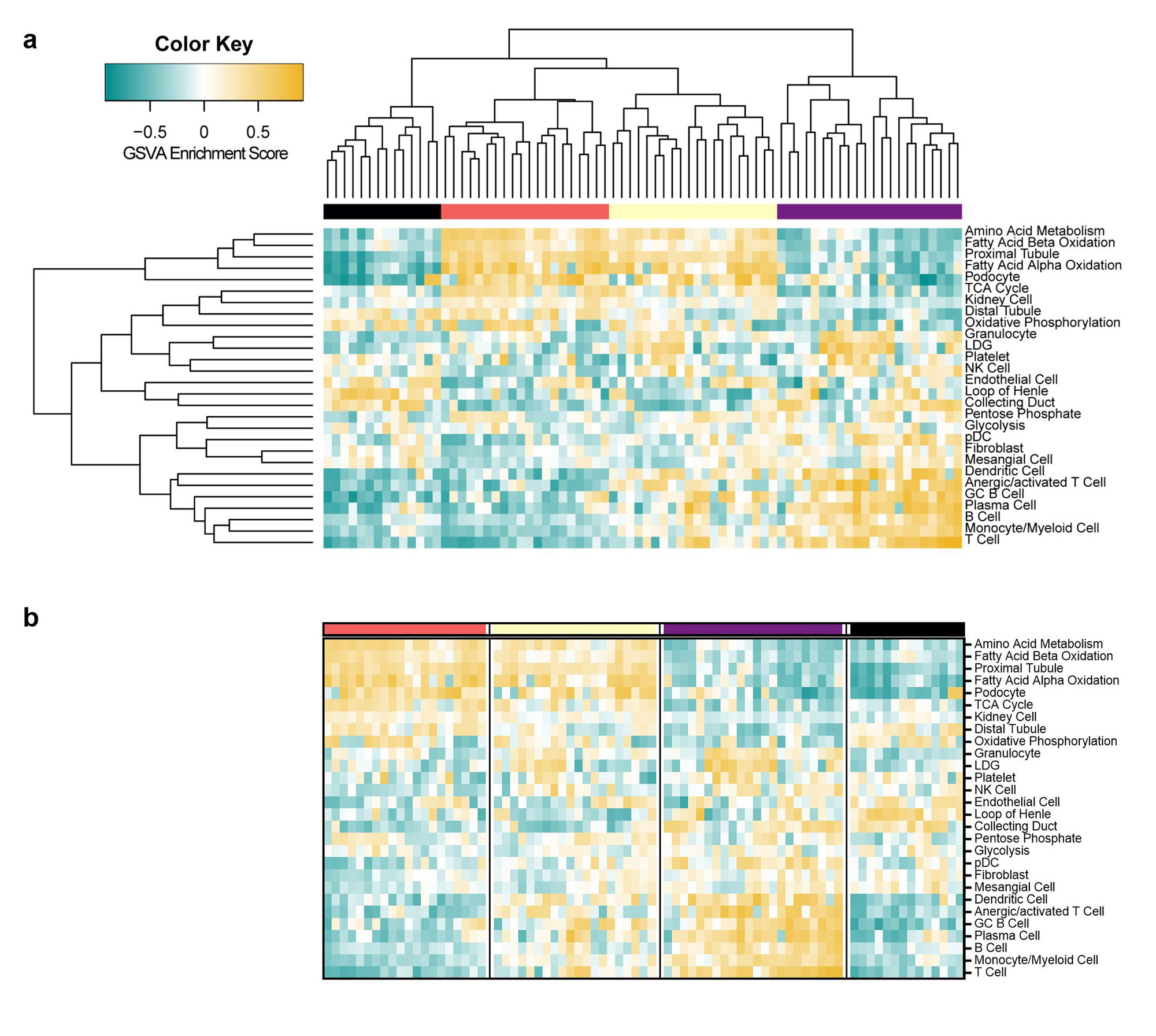
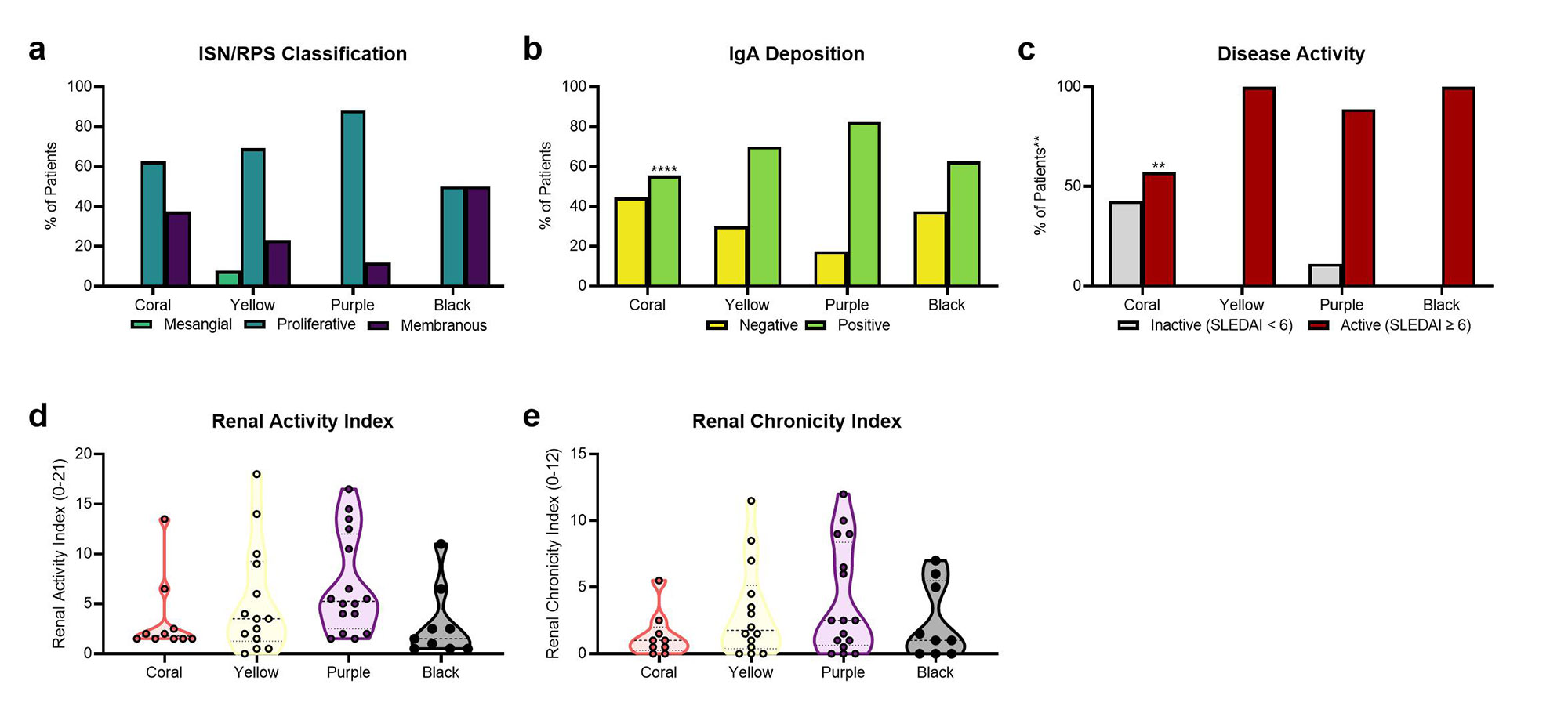
Results: Analysis of LN gene expression with GSVA revealed four distinct endotypes of LN (Fig. 1a), which could be ordered from least to most severe based on the profile of abnormal features (Fig. 1b). The least severe cluster (coral) exhibited minimal immune cell infiltrates or changes to kidney cell or metabolic signature expression. The second cluster (yellow) had increased expression of immune/inflammatory cell signatures, with minimal changes to kidney cell or metabolic signatures. The third cluster (purple) exhibited both increased expression of immune cell signatures and decreased expression of kidney cell and metabolic signatures. The final cluster (black) exhibited minimal immune cell gene expression, but decreased metabolism signatures and increased endothelial cell, fibroblast, and mesangial cell signatures. Of the LN samples with paired histology, 24/33 samples with proliferative LN were found in clusters with increased immune/inflammatory cell signatures (15/33 in the purple cluster, 9/33 in the yellow cluster) (Fig. 2a). Notably, the percentage of patients with IgA deposition on the glomerular basement membrane was highest in the purple cluster, and significantly lower in the coral cluster (Fig. 2b). Moreover, the percentage of patients with active disease determined by SLEDAI was lowest in the coral cluster (Fig. 2c). Mean renal activity and chronicity indices were not significantly different between clusters (Fig. 2d-e).
Conclusion: Transcriptomic analyses suggest distinct endotypes of LN, including: 1) minimal disease; 2) inflammatory disease without kidney cell damage or metabolic dysfunction; 3) inflammatory disease with kidney cell and metabolic dysfunction; and 4) markedly decreased kidney cell and metabolic function with little inflammation. Even though there is modest association with histologic phenotypes, the molecular endotypes suggest progression of LN from acute inflammatory to chronic dysfunctional kidney disease. The molecular endotypes of LN may be an important way to stage LN and provide useful information to guide therapy.
To cite this abstract in AMA style:
Kingsmore Allison K, Shrotri S, Bachali P, Shen N, Grammer A, Lipsky P. Transcriptomic Analysis of Lupus Nephritis Kidneys Identifies Molecular Endotypes [abstract]. Arthritis Rheumatol. 2022; 74 (suppl 9). https://acrabstracts.org/abstract/transcriptomic-analysis-of-lupus-nephritis-kidneys-identifies-molecular-endotypes/. Accessed .« Back to ACR Convergence 2022
ACR Meeting Abstracts - https://acrabstracts.org/abstract/transcriptomic-analysis-of-lupus-nephritis-kidneys-identifies-molecular-endotypes/