Session Information
Session Type: Abstract Submissions (ACR)
Background/Purpose
Mitochondria and DNA methylation play a main role in the development of Osteoarthritis. The aim of this work is to analyze the influence of the mitochondrial background on the DNA methylome of articular chondrocytes.
Methods
DNA methylation profiling was performed using the Infinium HumanMethylation27 beadchip. Previously, cartilage isolated DNA from 41 cartilage samples (13 from haplogorup J and 20 from haplogroup H) was bisulfite-modified using EZ DNA methylation kit and hybridized according to the manufacturerxs instructions. DNA methylation M-values were obtained and further compared between haplogroups using ANOVA and adjusting for cofounder effects of age, gender and disease status. Bonferroni post-hoc analysis was performed for analysing haplogroup pairwise differences. Enrichment in biological process and molecular function was tested by means of IPA (Ingenuity Pathway Analysis) software (Qiagen). All statistical analyses were conducted in R software.
Results
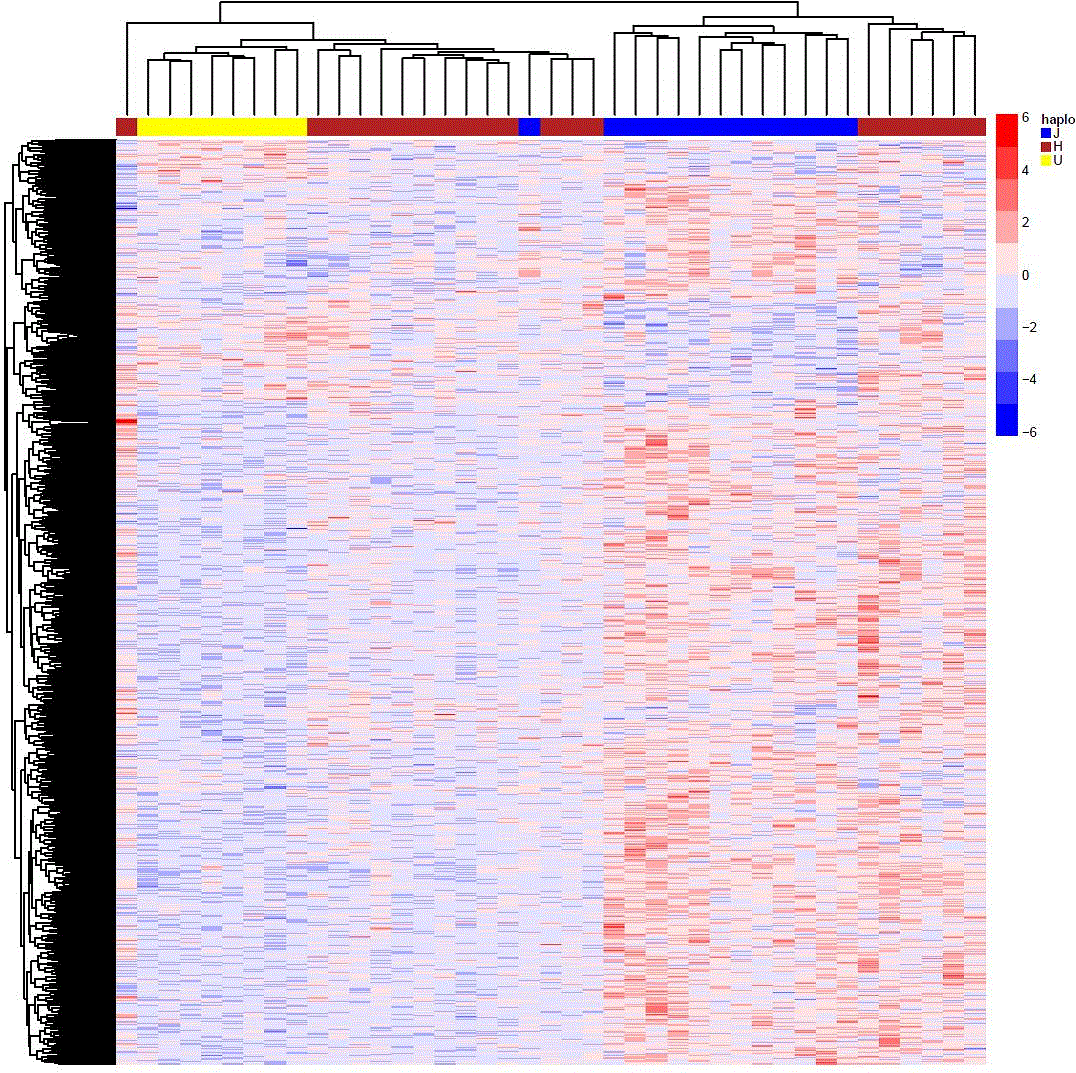
ANOVA analysis showed a total of 1926 CpG probes with a p-value under 0.05 (Figure 1); bonferroni post-hoc analysis allowed us to identify 538 significant probes (adjusted p-value < 0.05) between haplogroup H and haplogroup J; of these, 451 were more methylated in J and 87 were less methylated in J in relation to the most common haplogroup H.
A CpG site in the promoter region of fucosidase, alpha-L-1 (FUCA1) gene was the most differentially methylated probe between H and J groups, being the most methylated in haplogroup J; on the contrary, a CpG site located in the promoter region of homeobox D3 (HOXD3) gene, the second most differentially methylated probe between H and J groups, was the less methylated in haplogroup J, compared with the most common haplogroup H.
Among the most significant altered canonical pathways obtained with IPA software, those involving apoptosis signalling (p=0.013) and inflammatory response (p=0.0048) stand out. Besides, an enrichment of several pathways involved in TGF-β signalling (p=0.0107), iNOS signalling (p=0.0011), BMP signalling (p=0.0044), Protein kinase A signalling (p=0.0071), MAPK signalling (p=0.016) or PI3K signalling (p=0.0074) were also revealed. It is noteworthy that, to a greater or lesser extent, these pathways are involved in the OA process.
Conclusion
The mitochondrial genetic background seems to modify the nuclear epigenome of articular chondrocytes. Some of the altered pathways, mainly enhanced in carriers of the haplogroup H, have been previously described to be involved in the etiology of OA. The role played by the mtDNA haplogroups on Spanish patients with OA could be mediated by this particular epigenetic profile.
Figure 1. Heatmap showing the differential DNA methylome related to the mtDNA haplogroups
Disclosure:
I. Rego-Perez,
None;
J. Fernández-Tajes,
None;
M. Fernandez Moreno,
None;
A. Soto-Hermida,
None;
M. E. Vázquez-Mosquera,
None;
E. Cortés-Pereira,
None;
M. Tamayo,
None;
S. Relaño-Fernandez,
None;
A. Mosquera,
None;
N. Oreiro-Villar,
None;
C. Fernandez-Lopez,
None;
J. L. Fernandez,
None;
F. J. Blanco Garcia,
None.
« Back to 2014 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/the-mtdna-haplogroups-influence-the-dna-methylome-of-articular-chondrocytes/