Session Information
Date: Tuesday, November 10, 2015
Title: Spondylarthropathies and Psoriatic Arthritis Pathogenesis, Etiology Poster I
Session Type: ACR Poster Session C
Session Time: 9:00AM-11:00AM
Background/Purpose:
Reactive arthritis (ReA) is an inflammatory arthritis
that typically follows infection. Several agents microbial agents have been
implicated, particularly Shigella,
Salmonella, Campylobacter in the
gastrointestinal tract and Chlamydia trachomatis in the genitourinary tract.
However, the intestinal microbiome in these patients has never been characterized. The objective of this study was to
determine the community composition of pathogenic and commensal gut microorganisms
of ReA subjects from a highly prevalent region.
Methods: We
performed a prospective case-control study among patients with an infection
(urogenital or gastrointestinal) in the 3-6 months prior to enrollment (January
through October 2014). ReA was defined using the ASAS peripheral SpA criteria. Cases
were identified in the AGAR rheumatology clinic. Controls were recruited from
primary care clinics in the same hospital system in Guatemala. Fecal samples from 33 subjects with ReA and 31 controls were obtained and bacterial DNA was
extracted. 16S rDNA amplicons
targeting V4 region were sequenced using MiSeq (Illumina) to define the microbiota
composition. The obtained 16S rRNA sequences were
analyzed using the Quantitative Insights into Microbial Ecology (QIIME)
pipeline for analysis of community sequence data.
Results: There
were no significant differences in age, gender, HLA-B27 positivity or type of
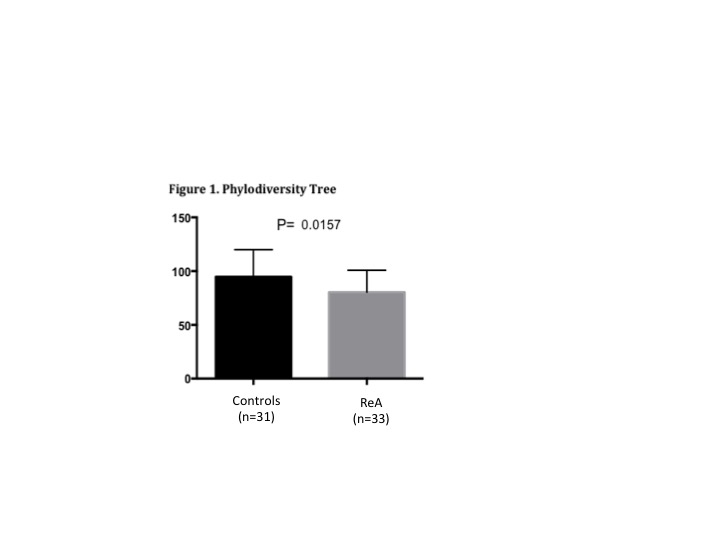
infections between the groups. 16S sequencing data showed that ReA patients had significantly less intestinal microbial
diversity compared to controls, as calculated by chao
(p<0.05) and phylogenetic tree (p<0.05; Figure 1). Taxonomic comparison
between groups was performed using LEfSe, which
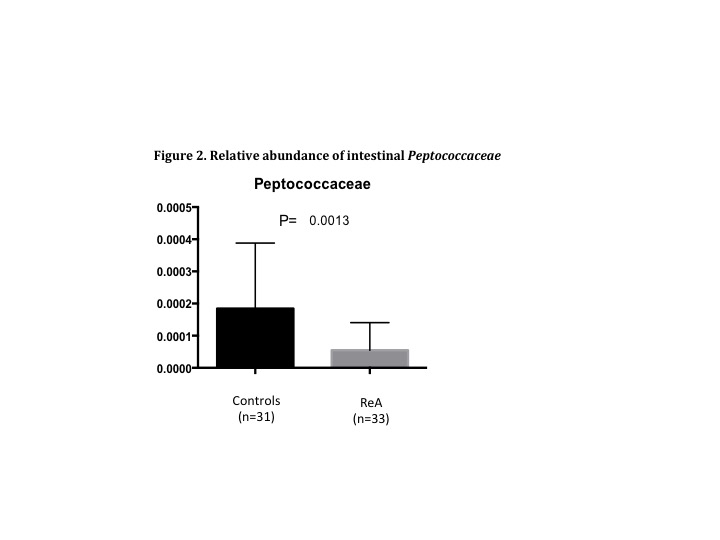
revealed a few significant differences (LDA score>2). While ReA microbiome samples were
enriched with Bifidobacteriaceae (p<0.05), there was a notable and significant decrease in intestinal Peptococcaceae (p<0.001; Figure 2). At the operational taxonomic unit
(OTU) level, there was a virtual absence of Paraprevotellaceae in the ReA
group (p<0.01).
Conclusion: Despite
the relatively small number of samples analyzed, the intestinal microbiome of ReA patients
appears to be less diverse than that of controls. Taxonomic differences were also
noted between groups, including the lower abundance of gut pathobionts
(i.e., Peptococcaceae).
Further evaluation of functional aspects of this microbiome
and its relationship to intestinal commensals and pathogens in various
patients’ phenotypes may provide further insights into its possible
contribution to ReA.
To cite this abstract in AMA style:
Ogdie-Beatty A, Ubeda C, Garcia Ferrer HR, Von Feldt J, Garcia Kutzbach A, Scher JU. The Microbiome of Reactive Arthritis in a Guatemalan Cohort [abstract]. Arthritis Rheumatol. 2015; 67 (suppl 10). https://acrabstracts.org/abstract/the-microbiome-of-reactive-arthritis-in-a-guatemalan-cohort/. Accessed .« Back to 2015 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/the-microbiome-of-reactive-arthritis-in-a-guatemalan-cohort/