Session Information
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: Systemic lupus erythematosus (SLE) is an autoimmune disease with a heterogeneous clinical picture. This study aimed to link genetic SLE predisposition with relevant clinical manifestations.
Methods: This study uses a discovery- and validation approach. First, we conducted an ICD-10 code-based search in the FinnGen consortium population database, which includes >218,000 individuals from Finland (data freeze 5) with extensive genetic and disease end-point information1. Through this search, we identified 30 datasets corresponding to the 11 ACR-82 classification criteria for SLE, containing cases with SLE-like manifestations such as polyarthritis, hemolytic anemia and immunological disturbances in serum. Summary statistics for 57 established SLE risk SNPs (p< 5×10-8 in the European population) with a validated cumulative effect on disease severity2 in SLE were retrieved for the selected datasets. Mendelian Randomization analysis using the inverse variance-weighted method was conducted to evaluate the effect of a high genetic SLE predisposition on each manifestation.
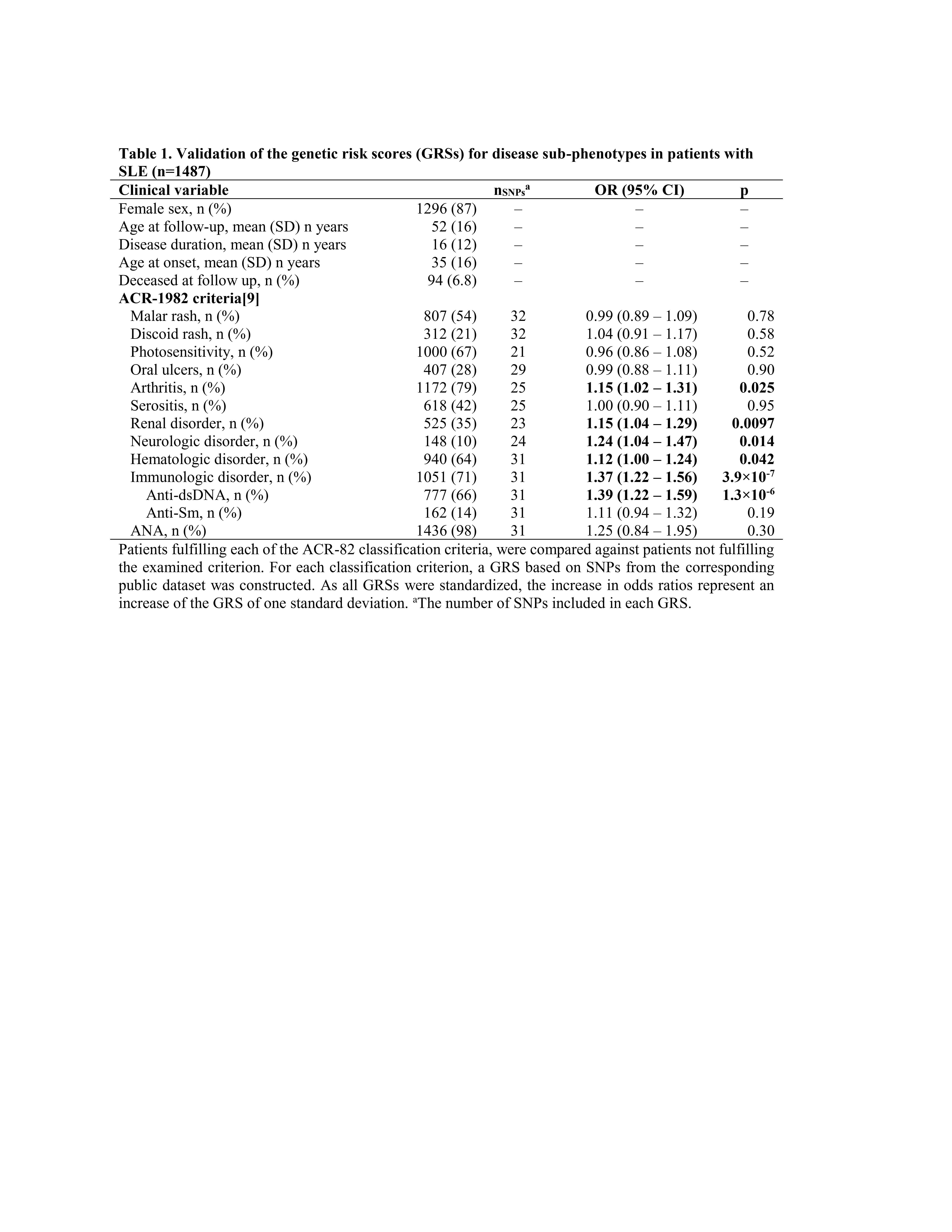
Second, validation was conducted in a clinical SLE cohort. Scandinavian patients with SLE (n=1487) who fulfilled ≥4 ACR-82/ACR-97/SLICC-2012 classification criteria were genotyped using the Immunochip or Global Screening Array (Illumina). Clinical data was collected from medical records. Standardized weighted genetic risk scores (GRSs) for each relevant manifestation were constructed for each patient based on the Finngen datasets. Associations between each GRS and the corresponding ACR-82 criterion in the clinical SLE cohort were evaluated using sex- and disease duration-adjusted logistic regression.
Results: In the FinnGen biobank, the cumulative effect of the 57 SLE risk SNPs was associated with an increased risk of rosacea, OR 1.09 (1.03–1.16), polyarthropathies, OR 1.10 (1.06–1.14), pleural effusions, OR 1.09 (1.04–1.14), and hemolytic anemia, OR 1.32 (1.10–1.58).
In the clinical cohort, 5 of the 11 GRSs generated from the FinnGen datasets were associated with their corresponding ACR-82 criterion: arthritis, OR 1.15 (1.02–1.31), renal disorder, OR 1.15 (1.04–1.29), neurologic disorder, OR 1.24 (1.04–1.47), hematologic disorder, OR 1.12 (1.00–1.24), and immunologic disorder, OR 1.37 (1.22–1.56), table 1.
Conclusion: By considering associations of SLE risk SNPs with SLE-like manifestations in the FinnGen biobank, construction and validation of GRSs for five of the eleven ACR-82 criteria was possible. The findings demonstrate known SLE risk gene variants to play a role in the development of at least half of the ACR-82 criteria, indicating a future possibility of using genetics to predict a variety of disease sub-phenotypes in SLE.
1Kurki, M.I., et al., FinnGen provides genetic insights from a well-phenotyped isolated population. Nature, 2023. 613(7944)
2Reid, S., et al., High genetic risk score is associated with early disease onset, damage accrual and decreased survival in systemic lupus erythematosus. Ann Rheum Dis, 2020. 79(3): p. 363-369.
To cite this abstract in AMA style:
Reid S, Sandling J, Pucholt P, Sayadi A, Frodlund M, Lerang K, Jönsen A, Sjowall C, Gunnarsson I, Syvänen A, Bengtsson A, Molberg Ø, Svenungsson E, Rudin A, Rantapaa-Dahlqvist S, Rönnblom L, Leonard D. The Construction and Validation of Sub-phenotype-specific Genetic Risk Scores in Systemic Lupus Erythematosus: A Novel Approach Using Large-scale Biobank Data [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/the-construction-and-validation-of-sub-phenotype-specific-genetic-risk-scores-in-systemic-lupus-erythematosus-a-novel-approach-using-large-scale-biobank-data/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/the-construction-and-validation-of-sub-phenotype-specific-genetic-risk-scores-in-systemic-lupus-erythematosus-a-novel-approach-using-large-scale-biobank-data/