Session Information
Session Type: Abstract Session
Session Time: 4:00PM-5:30PM
Background/Purpose: Still’s disease (systemic juvenile idiopathic arthritis in children, adult-onset Still’s disease in adults) is an enigmatic inflammatory condition that affects people of all ages. It is characterized by inflammasome activation and pyroptosis, and is often punctuated by striking elevation of IL-18. Increased interferon (IFN) signaling, which promotes pyroptosis, has been described in Still’s disease complicated by either macrophage activation syndrome (MAS) or an emergent form of lung disease (LD) with drug-induced hypersensitivity reaction (DHR), but it is unclear whether IFN signaling is an active participant or is epiphenomenal. Here, we explore the relationship between genetic variation and IFN signaling in Still’s disease.
Methods: We examined consecutive Still’s disease patients enrolled in an IRB approved research study at the NIH Clinical Center. Whole blood expression of 28 IFN-stimulated genes (ISG) was quantified with custom NanoString arrays and expressed as a normalized score. Trio whole exome sequencing (WES) was performed in 41 probands and candidate causative variant lists (protein-altering de novo, recessive, or X-linked variants of canonical transcripts; popfreqmax frequency < 0.01) were generated. Lists of genes containing candidate causative variants were compiled for individuals with the highest or lowest ISG scores and subjected to pathway over-representation analysis (ORA) and network topology analysis (NTA) using the Web-based Gene Set Analysis Toolkit (webgestalt.org).
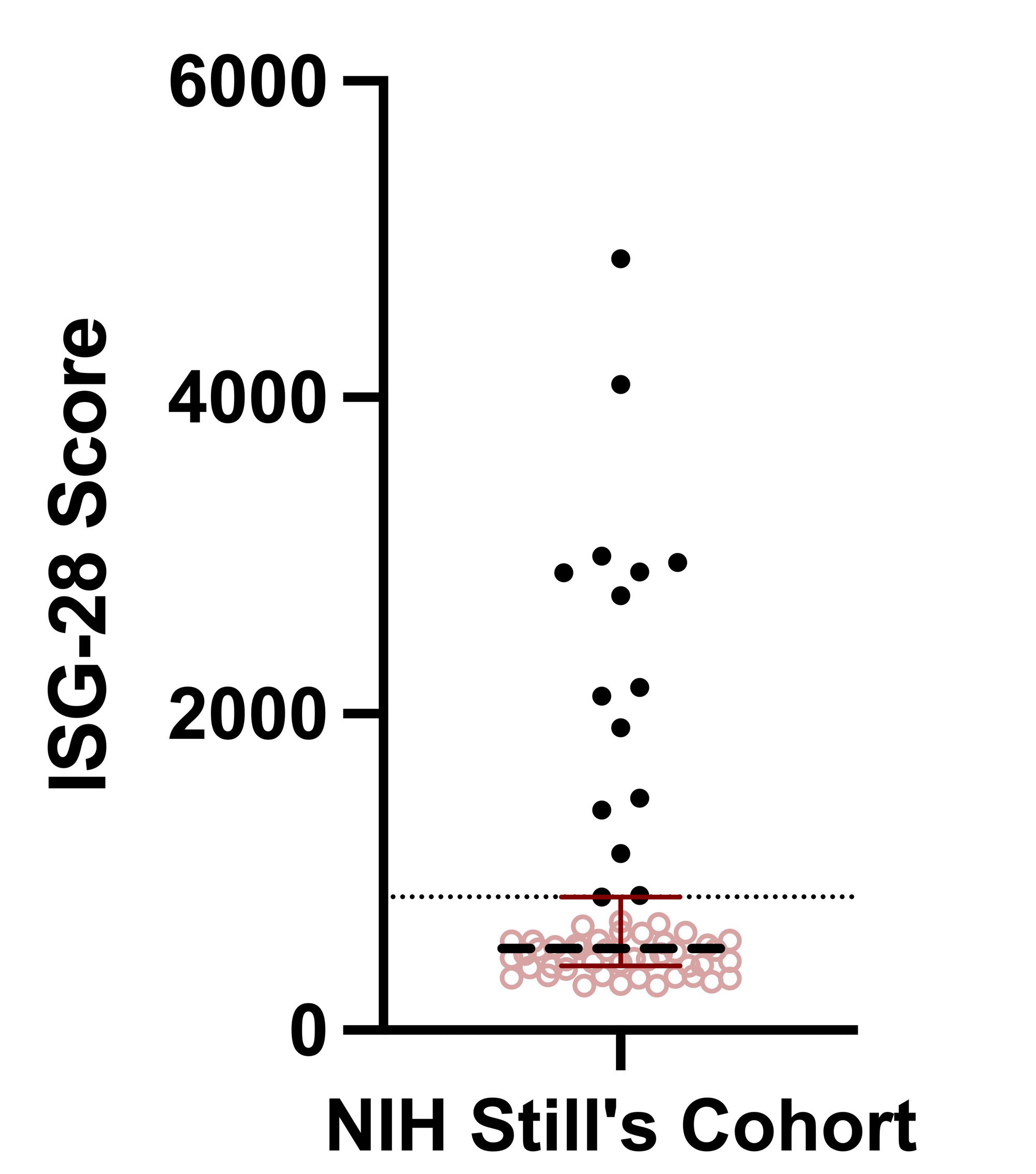
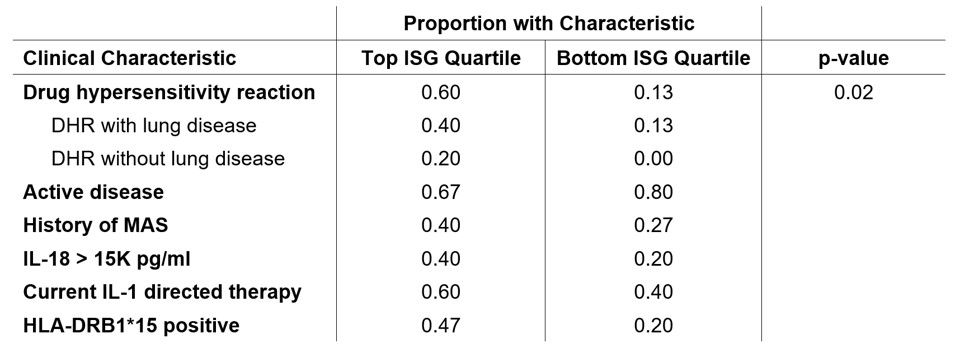
Results: ISG scores among 59 Still’s patients were negatively skewed and the upper quartile included 15 people (Figure 1). DHR was more common in the top quartile than in the bottom quartile (0.6, 0.13; p = 0.02), while rates of active disease, IL-18 > 15K, use of IL-1 directed therapy, HLA-DRB1*15 carriage and history of MAS did not differ between the groups (Table 1). Trio WES identified 83 candidate genes among the top ISG quartile. ORA of these 83 candidate genes revealed enrichment in the macrophage activation (ratio = 21.7, p = 3.4E-7, FDR = 0.0055), type I IFN biosynthetic process (ratio = 77.9, p = 6.7E-6, FDR = 0.03) and myeloid leukocyte activation (ratio = 5.0, p = 1.9E-5, FDR = 0.039) pathways. Most people in the top ISG quartile (11/15) had candidate causative variation within these 3 pathways. Chemokine production, regulation of IL-18 production, and NF-KB signaling also had evidence of over-representation (p < 0.0005, FDR > 0.05). NTA also demonstrated enriched pathways, including the MyD88-dependent TLR signaling pathway (corrected p = 0.0045). ORA/NTA of 81 candidate genes from 15 subjects with the lowest ISG scores did not reveal enrichment of any pathway.
Conclusion: In our Still’s cohort, high ISG score was associated with the presence of DHR but not MAS. Unbiased analysis of trio WES revealed that people with high ISG scores had specific enrichment of potentially causative variation in overlapping innate immune pathways, notably type I IFN production, MyD88/TLR signaling, and macrophage/myeloid leukocyte activation. This suggests that rare de novo or recessive variation contributes to the pathophysiology of Still’s disease, where it promotes enhanced IFN signaling and may predispose to LD/DHR.
To cite this abstract in AMA style:
Correia Marques M, Deng Z, Chowdhury N, Schmitz E, Platukus A, Brooks S, Lake C, Bergeron L, Millwood M, Ombrello M. Still’s Disease Patients with High Interferon-stimulated Gene Expression Have Enrichment of Rare, de Novo and Recessive Protein Altering Variants in Innate Immune Pathways [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/stills-disease-patients-with-high-interferon-stimulated-gene-expression-have-enrichment-of-rare-de-novo-and-recessive-protein-altering-variants-in-innate-immune-pathways/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/stills-disease-patients-with-high-interferon-stimulated-gene-expression-have-enrichment-of-rare-de-novo-and-recessive-protein-altering-variants-in-innate-immune-pathways/