Session Information
Session Type: Poster Session C
Session Time: 1:00PM-3:00PM
Background/Purpose: The skin is recognized as a window into the immunopathogenic mechanisms driving the vast phenotypic spectrum of psoriatic disease.
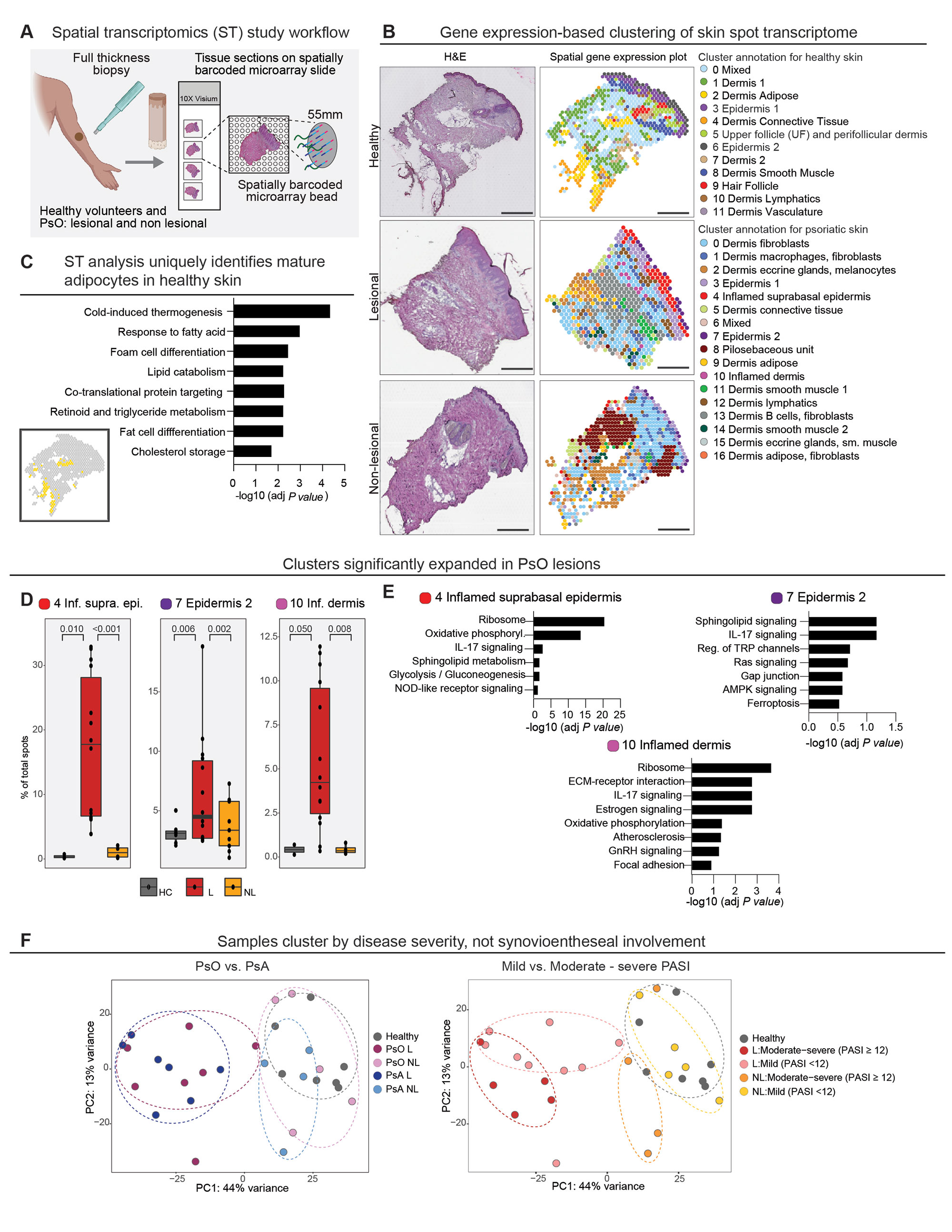
Methods: To better decipher the cellular landscape of both healthy and psoriatic skin, we employed spatial transcriptomics (ST), a ground-breaking technology that precisely maps gene expression from histologically-intact tissue sections (Fig. 1A).
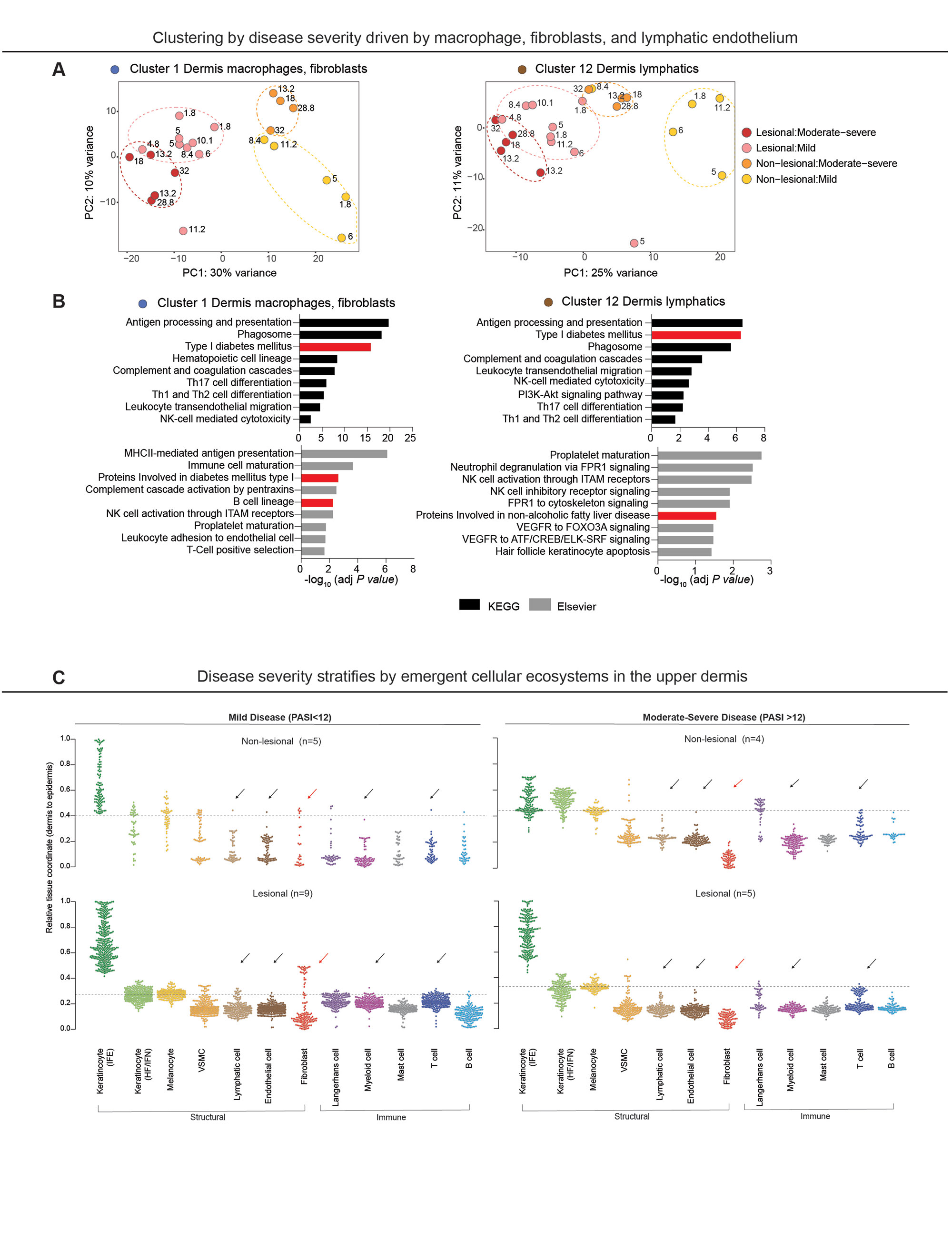
Results: Findings gleaned from computationally integrating our 23 matched lesional and non-lesional psoriatic and 7 healthy control samples with publicly-available single-cell ribonucleic acid (RNA) sequencing datasets established the ability of ST to recapitulate the tissue architecture of both healthy and inflamed skin (Fig. 1B) and highlighted topographic shifts in the immune cell milieu, from a predominantly perifollicular distribution in steady-state skin to the papillary and upper reticular dermis in psoriatic lesional skin. We also incidentally discovered that ST’s ability to ascertain gene expression patterns from intact tissue rendered it particularly conducive to studying the transcriptome of lipid-laden cells such as dermal adipose tissue and sebaceous glands (Fig. 1C), whose expression profiles are typically lost in the process of tissue handling and dissociation for bulk and single-cell RNA seq. Unbiased clustering of pooled healthy and psoriatic samples identified two epidermal clusters and one dermal cluster that were differentially expanded in psoriatic lesional skin (p values ≤0.05) (Fig. 1D); pathway analysis of these clusters revealed enrichment of known psoriatic inflammatory pathways (Fig. 1E). Unsupervised classification of skin-limited psoriasis and psoriatic arthritis samples revealed stratification by cutaneous disease severity or Psoriasis Area and Severity Index (PASI) score and not by presence or absence of concomitant systemic/synovial disease (Fig. 1F). Remarkably, this PASI-dependent segregation was also evident in distal, non-lesional samples and was driven by the dermal macrophage and fibroblast cluster and the lymphatic endothelium (Fig. 2A). Inquiry into the mechanistic drivers of this observed stratification yielded enrichment of pathways associated with key T cell and innate immune cell activation, B cells, and metabolic dysfunction (Fig. 2B). Finally, tissue scale computational cartography of gene expression revealed differences in regional enrichment of specific cell types across phenotypic groups, most notably upward extension of fibroblasts to the upper dermis in both lesional and non-lesional samples from mild psoriasis and restriction to the lower dermis in the moderate-to-severe psoriasis samples (Fig. 2C), suggesting that disease severity stratification may be driven by emergent cellular ecosystems in the upper dermis.
Conclusion: Thus, we have been able to successfully leverage ST integrated with independently-generated single-cell RNA seq data to spatially define the emergent cellular ecosystems of healthy and matched psoriatic lesional and non-lesional skin and in so doing, demonstrated the value of ST in unearthing the genetic groundwork at both the site of inflammation and in distal, clinically-uninvolved skin.
To cite this abstract in AMA style:
Castillo R, Sidhu I, Dolgalev I, Subudhi I, Yan D, Konieczny P, Hsieh B, Chu T, Haberman R, Selvaraj S, Shiomi T, Medina R, Vasudevanpillai Girija P, Heguy A, Loomis C, Chiriboga L, Meehan S, Ritchlin C, Garcia-Hernandez M, Carucci J, Neimann A, Naik S, Scher J. Spatial Transcriptomics Stratifies Health and Psoriatic Disease Severity by Emergent Cellular Ecosystems [abstract]. Arthritis Rheumatol. 2022; 74 (suppl 9). https://acrabstracts.org/abstract/spatial-transcriptomics-stratifies-health-and-psoriatic-disease-severity-by-emergent-cellular-ecosystems/. Accessed .« Back to ACR Convergence 2022
ACR Meeting Abstracts - https://acrabstracts.org/abstract/spatial-transcriptomics-stratifies-health-and-psoriatic-disease-severity-by-emergent-cellular-ecosystems/