Session Information
Session Type: Abstract Session
Session Time: 1:00PM-2:30PM
Background/Purpose: Sjögren’s disease (SjD) is an autoimmune disorder characterized by inflammatory destruction of the exocrine glands. SjD heterogeneity has been described through the presence or absence of the anti-SSA/Ro autoantibody. Previous studies examining whole blood of SjD patients reported transcriptional differences between SSA/Ro+ and SSA/Ro– subtypes. Here, we leveraged single-cell transcriptomics in peripheral blood mononuclear cells (PBMCs) to refine our understanding of cell type-specific variation in pathogenic pathways across different subpopulations of SjD patients.
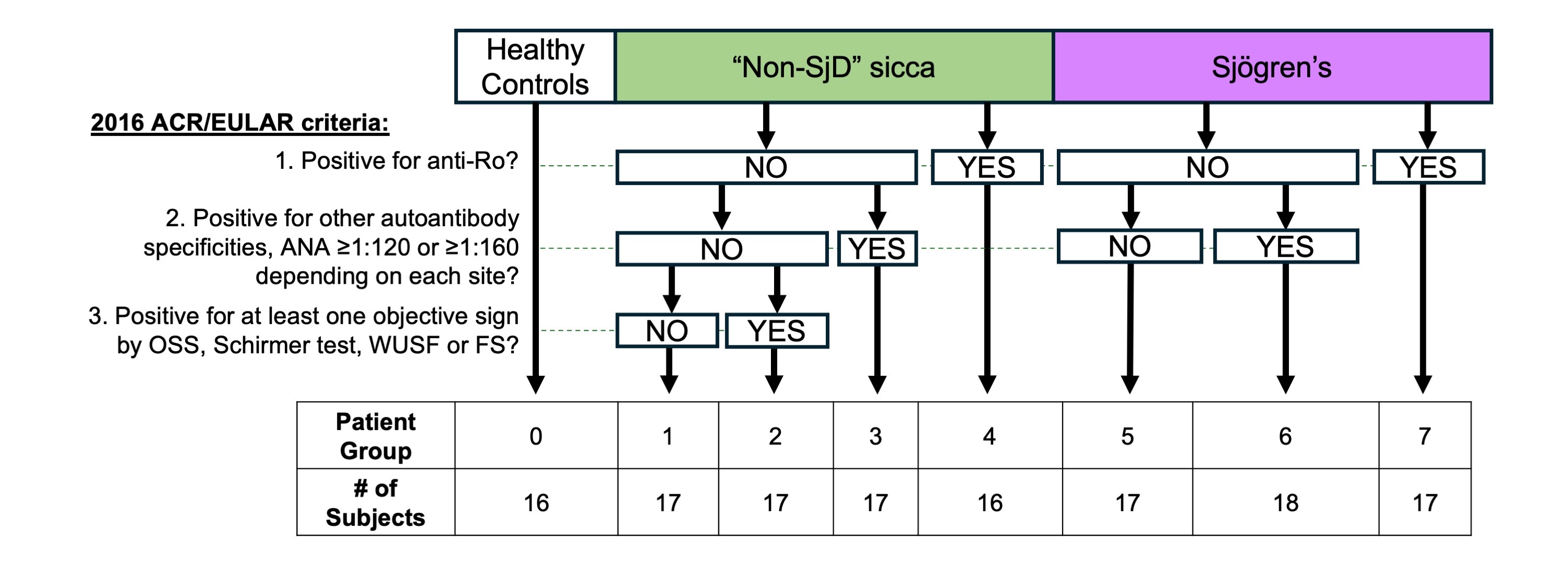
Methods: PBMCs were isolated from human subjects with SjD, subjects with sicca symptoms not meeting SjD criteria, and healthy controls. Subjects were stratified into Groups 0-7 (Fig.1). 10X Genomics Chromium Single Cell RNA-sequencing was performed and sequencing data were demultiplexed using the 10X Genomics Cell Ranger (v 7.0.0) pipeline and aligned to the GRCh38 human genome. After quality control, 789,780 cells were integrated and clustered using Seurat1. Differential expression (DE) analyses were performed using NEBULA2. Significantly DE transcripts were input into Ingenuity Pathway Analysis (IPA).
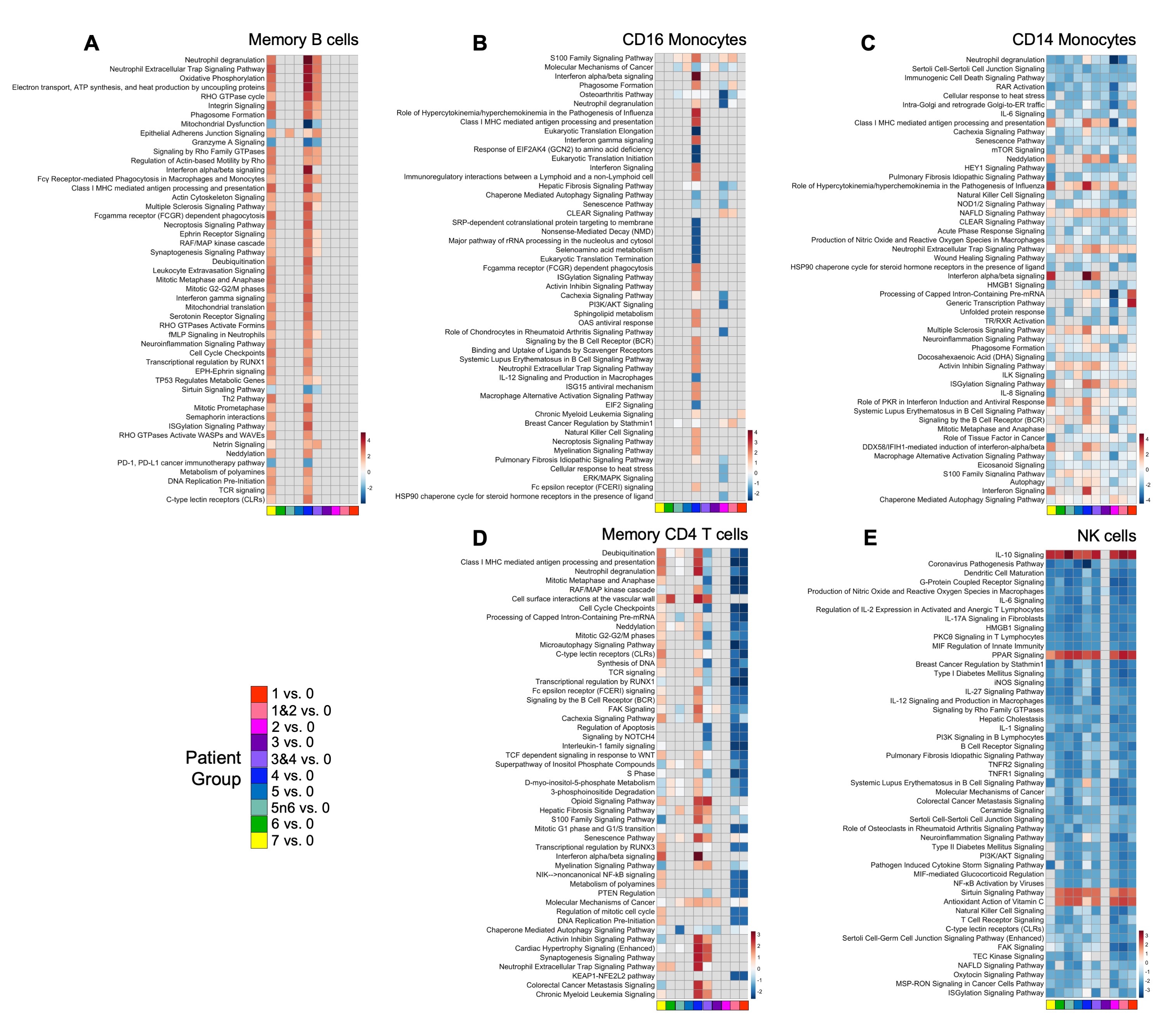
Results: Our analyses confirmed previously reported overexpression of type 1 interferon (T1IFN)-related genes in the anti-SSA/Ro+ subset of SjD patients (group 7). Upregulation of T1IFN signaling was also present in patients who were anti-SSA/Ro+ but did not meet SjD criteria (group 4). In memory B cells, many pathways involving energy production, cellular proliferation, and cell migration were upregulated in groups 4 and 7 (Fig. 2A). Dysregulation of CD16 monocytes was primarily seen in groups 2, 4, and 5. An increase in S100 signaling was also observed in these groups. Group 4 displayed a unique CD16 monocyte transcriptional profile distinguished by increased activity in immunoregulatory pathways and decreased activity in chaperone-mediated autophagy and translation-coupled pathways (Fig. 2B). A different pattern emerged in CD14 monocytes – increased autophagy in group 4 but decreased autophagy in group 7. Outside of autophagy, CD14 monocytes showed similar trends in downregulation of common pathways in groups 4 and 7: IL-6, mTOR, and RAR activation among others (Fig. 2C). Most patient groups showed similar trends in NK cell dysregulation, where IL-10 and PPAR signaling were upregulated while IL-1, IL-12, IL-27, and many other pathways were downregulated (Fig. 2E).
Conclusion: Immune cells demonstrate cell type-specific dysregulation in key pathways governing the differentiation of unique cell types and regulation of immune responses, both in patients who meet SjD criteria and those with non-SjD sicca. Further analyses of shared underlying processes could help elucidate molecular predictors of the transition from early symptoms to confirmed disease.
References: 1. Satija, R. et al. Spatial reconstruction of single-cell gene expression data. Nature Biotechnology 33, 495-502 (2015); 2. He, L. et al. NEBULA is a fast negative binomial mixed model for differential or co-expression analysis of large-scale multi-subject single-cell data. Commun Biol 4, 629 (2021).
To cite this abstract in AMA style:
Li J, Li C, Khatri B, Stolarczyk A, Tessneer K, Rasmussen A, Guthridge J, James J, Scofield R, Li H, Farris A, Sivils K, Lessard C. Sjögren’s Disease and Non-Sjögren’s Sicca Patient Subsets Exhibit Cell Type-specific Transcriptional Dysregulations That May Identify Early Molecular Predictors of Disease Transition [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/sjogrens-disease-and-non-sjogrens-sicca-patient-subsets-exhibit-cell-type-specific-transcriptional-dysregulations-that-may-identify-early-molecular-predictors-of-disease-transition/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/sjogrens-disease-and-non-sjogrens-sicca-patient-subsets-exhibit-cell-type-specific-transcriptional-dysregulations-that-may-identify-early-molecular-predictors-of-disease-transition/