Session Information
Date: Friday, November 6, 2020
Title: Innate Immunity Poster
Session Type: Poster Session A
Session Time: 9:00AM-11:00AM
Background/Purpose: Monocytes are pivotal producers of key inflammatory cytokines that drive autoinflammatory diseases. In SAVI, constitutive STING activation causes chronic activation with increased type-I IFN response gene expression in whole blood, and in NOMID, constitutive activation of the NLRP3 inflammasome leads to IL-1β mediated chronic inflammation. The effects of the respective activation of STING or the NLRP3 inflammasome on monocyte activation and differentiation are poorly understood. We used single cell RNA sequencing (scRNAseq) to profile purified monocytes from 2 SAVI and 3 NOMID patients.
Methods: Monocytes were positively selected from peripheral blood using CD14+ MicroBeads (Miltenyi). Transcription profiles from bead-purified monocytes from 2 SAVI and 3 NOMID patients (pts) enrolled in an IRB approved protocol (NCT02974595) were generated; 3000 monocytes were prepared for scRNAseq using Chromium™ Single Cell 3’ v2 Reagent Kit (10x genomics) and were sequenced (NextSeq®, Illumina) targeting 60,000 reads/cell. Files were demultiplexed and aligned to the GRCh38 transcriptome reference using Cell Ranger. Gene-barcode-matrices that passed quality control were analyzed individually and integrated; monocyte subsets were identified by a shared nearest neighbor (SNN) modularity optimization-based clustering algorithm (Seurat). Cell subsets were annotated based on cell-type specific marker expression. Each cell is also annotated as a classical (CM), intermediate (IM) or non-classical (NCM) monocyte (SingleR using MonacoImmuneData reference).
Results: Gene expression of CD14 and FCGR3A/CD16 transcripts and SNN modularity optimization-based clustering algorithms identified 6 monocyte subsets for all samples (active SAVI (n=2), active NOMID (n=2) and inactive NOMID (n=2)) (Fig.1 A, B). In SAVI pts, CD16+ NCM and CD14+CD16+ IM are expanded. Interestingly, in SAVI all monocyte subsets strongly express IFNB1 with less IL1B expression. CX3CR1 is high in CD16 NCM subsets. In contrast, in NOMID pts, CD14+ CM subpopulations are expanded. NOMID monocytes do not express IFNB1 and IL1B expression was stronger in CD14+ CM from the 2 inactive than active NOMID pts. Comparing active and inactive NOMID, the percentage of CD14+ cells decreases as well as their CCR2 expression but not CCR1 expression. Furthermore, CX3CR1 expression drops in CD16+ NCM in inactive NOMID (Fig. 2).
Conclusion: In the active, NLRP3-inflammasome mediated disease, NOMID, classical monocytes (CM) subsets are expanded while in the Type-1 interferonopathy, SAVI, CD16+ NCM are expanded. The distinct expression of IFNB1 transcripts in SAVI only and the chemokine receptor modulation in active (SAVI and NOMID) and inactive (NOMID) disease, suggest disease-specific activation dependent monocyte subset heterogeneity in chronic autoinflammatory diseases. Gene expression profiling is ongoing and may contribute to the characterization of the monocyte diversity in NOMID and SAVI that propagates chronic inflammation and tissue damage.
Funding for this study was provided in part by the Division of Intramural Research, NIAID and NHLBI, NIH.
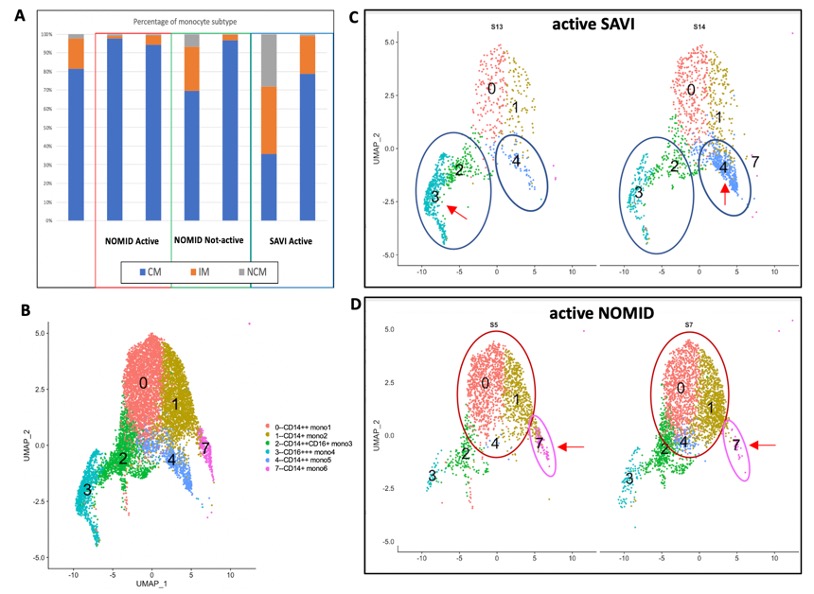 Figure 1. A. Each cell is annotated to be a classical (CM), intermediate (IM) or non-classical monocyte (NCM) by SingleR using MonacoImmuneData reference. Expansion of IM and NCM populations in one SAVI patient S13 and expansion of CM populations in mostly active NOMID patients and a patient who has been treated for 3 months with anakinra were quantified. B. Six monocyte subsets were identified by a shared nearest neighbor (SNN) modularity optimization- based clustering algorithm using Seurat. C, D. In active SAVI CD16+ mono3 (2), mono4 (3) and CD14+ mono5 (4) subsets were expanded, while in active NOMID expansion of CD14+ subsets mono1 (0), mono2 (1) and mono6 (7) were seen.
Figure 1. A. Each cell is annotated to be a classical (CM), intermediate (IM) or non-classical monocyte (NCM) by SingleR using MonacoImmuneData reference. Expansion of IM and NCM populations in one SAVI patient S13 and expansion of CM populations in mostly active NOMID patients and a patient who has been treated for 3 months with anakinra were quantified. B. Six monocyte subsets were identified by a shared nearest neighbor (SNN) modularity optimization- based clustering algorithm using Seurat. C, D. In active SAVI CD16+ mono3 (2), mono4 (3) and CD14+ mono5 (4) subsets were expanded, while in active NOMID expansion of CD14+ subsets mono1 (0), mono2 (1) and mono6 (7) were seen.
 Figure 2. Selected gene expression patterns in SAVI and NOMID monocytes. In SAVI, active patients’ monocytes have strong IFNB1 and moderate IL1B expression. CD16+ monocytes from active SAVI and NOMID pts. express CX3CR1 transcripts. NOMID patients do not express IFNB1. In inactive NOMID patients’ monocytes express more IL1B but lower CCR2 transcript levels than in clinically active NOMID patients.
Figure 2. Selected gene expression patterns in SAVI and NOMID monocytes. In SAVI, active patients’ monocytes have strong IFNB1 and moderate IL1B expression. CD16+ monocytes from active SAVI and NOMID pts. express CX3CR1 transcripts. NOMID patients do not express IFNB1. In inactive NOMID patients’ monocytes express more IL1B but lower CCR2 transcript levels than in clinically active NOMID patients.
To cite this abstract in AMA style:
Zhang Y, Marrero B, de Jesus A, Alehashemi S, Chen J, Shi R, Zhou H, Dalgard C, Boehm M, Goldbach-Mansky R. Single Cell RNA-seq to Characterize Monocyte Subtypes in the Autoinflammatory Interferonopathy, SAVI and the Inflammasomopathy, NOMID [abstract]. Arthritis Rheumatol. 2020; 72 (suppl 10). https://acrabstracts.org/abstract/single-cell-rna-seq-to-characterize-monocyte-subtypes-in-the-autoinflammatory-interferonopathy-savi-and-the-inflammasomopathy-nomid/. Accessed .« Back to ACR Convergence 2020
ACR Meeting Abstracts - https://acrabstracts.org/abstract/single-cell-rna-seq-to-characterize-monocyte-subtypes-in-the-autoinflammatory-interferonopathy-savi-and-the-inflammasomopathy-nomid/
