Session Information
Session Type: Poster Session B
Session Time: 9:00AM-11:00AM
Background/Purpose: Several groups of individuals are at higher risk for SLE, including those with African American ancestry (AA), lupus-associated autoantibodies (ANA+), or some clinical symptoms of SLE, termed incomplete lupus erythematosus (ILE). However, only a minority of ANA+ individuals develop autoimmune disease, and most ILE patients never progress to SLE. Metabolic pathways are known to be involved in myeloid cell dysregulation; however, whether they contribute to the progression of autoimmune disease remains unclear. This study determined whether alterations in myeloid and B cell population frequencies or activation of particular cellular processes are dysregulated in subjects at-risk of SLE.
Methods: PBMCs from 32 subjects from African (AA) or European ancestry (EA), divided evenly among healthy individuals (ANA-), healthy with autoantibodies (ANA+), ILE, and SLE. We performed multi-omic single-cell experiments (5′ scRNA-seq/137-plex CITE-seq and BCR/TCR repertoire analysis) to assay for distinct disease-associated clusters, differential gene signatures and dysregulated pathways.
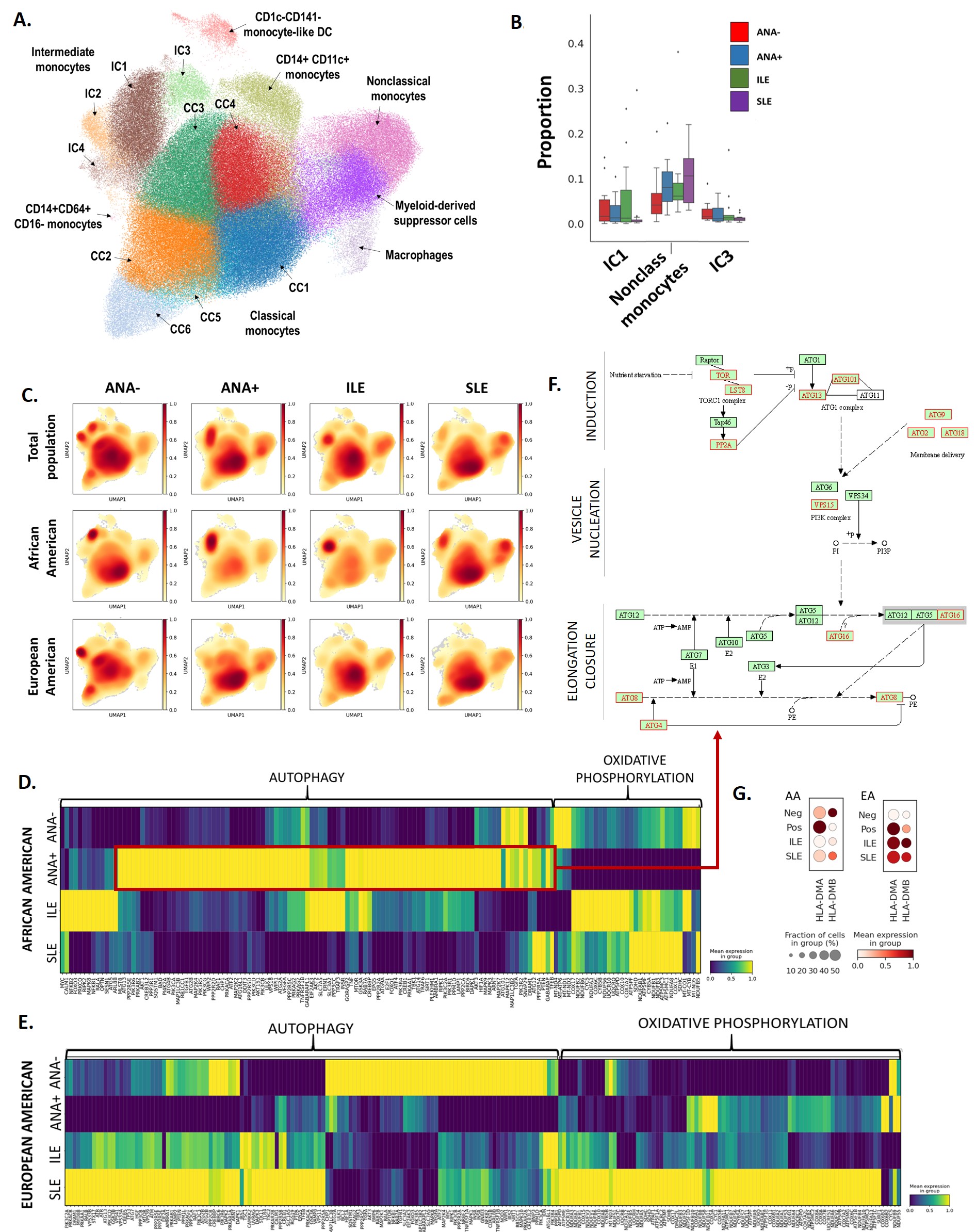
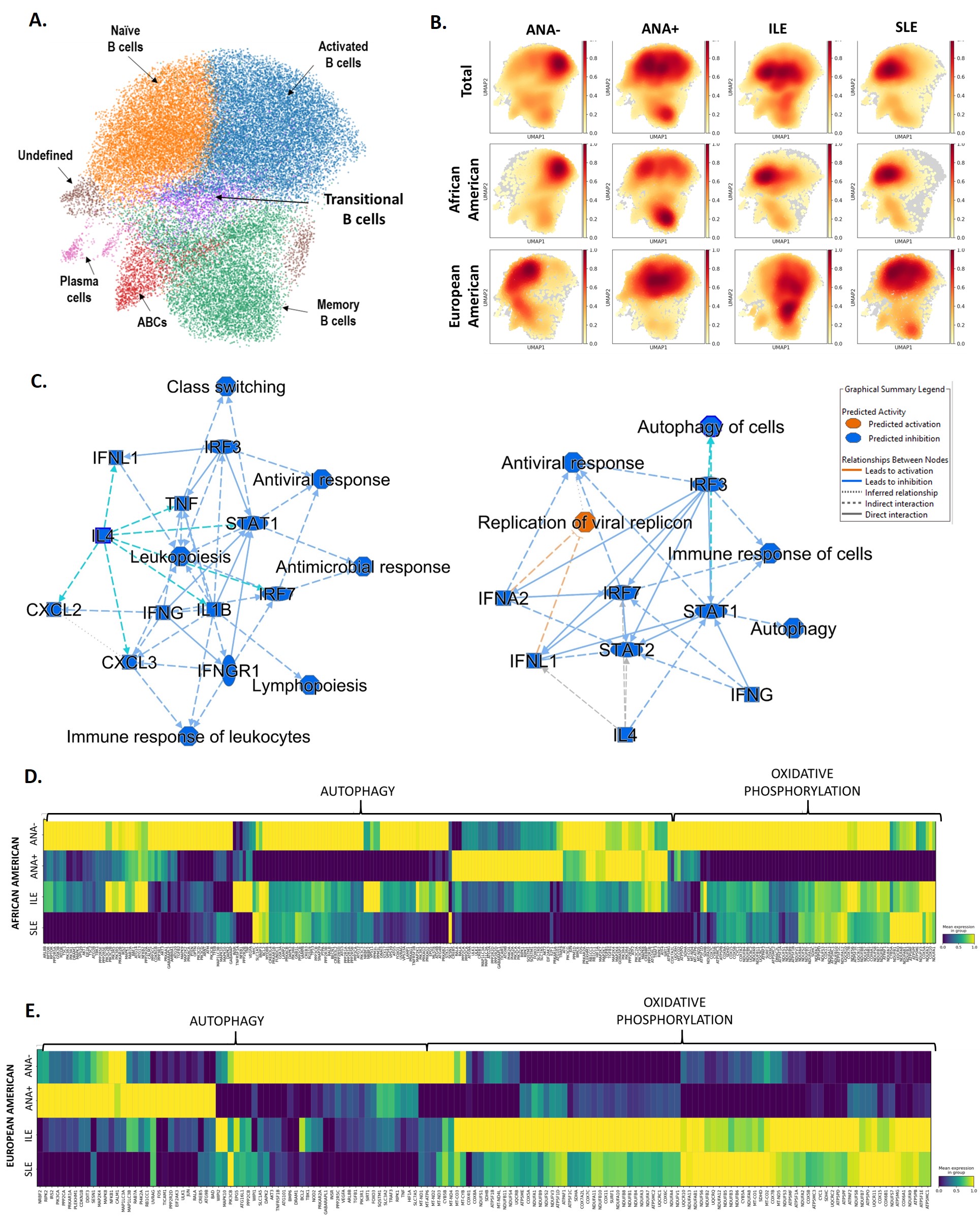
Results: We obtained profiles for 324,721 cells across all PBMCs. We identified 8 distinct myeloid (Fig 1A) and 7 B cell clusters across all subjects (Fig 2A). The proportion of cells in each cluster varied by disease group and ancestry (Fig 1B,C, 2B). The proportion of nonclassical monocytes was higher in SLE with variations between ANA+ and ILE (Fig 1B,C). Intermediate monocyte fractions were lower in SLE (Fig 1B,C). Fractions of naïve B cells were already higher in EA ANA-, while memory B cells are higher in AA ANA+ and EA ILE, EA SLE (Fig. 2B). Analysis of differentially expressed genes revealed the importance of metabolic processes in both cell types, such as autophagy and oxidative phosphorylation that varied between ancestries (Fig 1D-E, 2D-E). A discrete part of autophagy pathway is upregulated in myeloid cells of AA ANA+, while a different set of genes is upregulated in AA ILE (Fig 1D). Autophagy profile is detected earlier in EA; its progression is observed along disease transition and cell states (Fig 1E). Specific autophagy-related genes are already upregulated in EA ANA-. Oxidative phosphorylation pathway is downregulated in AA ANA+ but upregulated in AA ILE and AA SLE (Fig 1D). In EA, upregulation was mainly observed in SLE patients (Fig 1E). Involvement of antigen presentation pathways and altered expression of HLA-DM were detected in AA, which might be related to autophagy dysregulation (Fig 1F, G). Oxidative phosphorylation pathway is upregulated in B cells of EA ILE and EA SLE (Fig. 2E), however it is downregulated in AA ANA+ (Fig. 2D). We found additional signatures in B cells suggestive of the activation of pathways related to antiviral response, class switching and lymphopoiesis, as well as the involvement of transcription factors (IFN, TNF, IL4, STAT) in SLE (Fig.2C).
Conclusion: Dysregulation of oxidative phosphorylation and autophagy pathways in both monocyte and B cell populations is suggestive of a preclinical autoimmunity development trajectory. Alterations of these processes may vary by ancestral background, reflecting the heterogeneity of SLE presentation and trajectory.
To cite this abstract in AMA style:
Bylinska A, Smith M, Slight-Webb S, Guthridge C, Marlin C, Thomas K, Wright C, Beel M, Macwana S, DeJager W, James J, Guthridge J. Single-cell Multi-Omic Evaluation of Differences in Immune Cell Populations in Progression Toward Systemic Lupus Erythematosus [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/single-cell-multi-omic-evaluation-of-differences-in-immune-cell-populations-in-progression-toward-systemic-lupus-erythematosus/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/single-cell-multi-omic-evaluation-of-differences-in-immune-cell-populations-in-progression-toward-systemic-lupus-erythematosus/