Session Information
Session Type: Poster Session (Monday)
Session Time: 9:00AM-11:00AM
Background/Purpose: Studies examining intracellular micro-RNA (miR) expression in PBMCs of patients with Systemic Lupus Erythematous (SLE) have identified distinct disease-associated changes in expression. We have previously demonstrated that estrogen lowers the threshold of immune cell activation to a greater extent in females and leads to significantly enhanced toll-like receptor (TLR)7 and TLR8 expression. Moreover, our studies have also shown enhanced estrogenic responses in PBMCs from SLE patients compared to healthy controls. Furthermore, our recent data demonstrates estrogen-mediated upregulation of extracellular vesicle (EV) production via enhanced TLR8 expression in THP-1 cell lines and unique miR signatures in EVs isolated from SLE patient plasma and urine. The overall objective of the current study is to identify novel estrogen-regulated mRNAs and miRs that may be responsible, in part, for the unique miR signatures observed in EVs from SLE patients.
Methods: Our cohort for this study consisted of five post-adolescent, premenopausal, female donors. Human PBMCs were isolated from whole blood and cultured in hormone free conditions before stimulation with 10 nM of 17β-estradiol (estrogen; E2) for 24 hr. E2-treated samples were compared to untreated controls for each donor. RNA was isolated from cell lysates and RNA libraries were prepared for RNA-sequencing (RNAseq). Global RNA reads were analyzed and cross-referenced with a sequencing database (miRBase) of known miRs and mRNAs. Results for mRNA and miR data were analyzed collectively on Ingenuity Pathway Analysis (IPA) software.
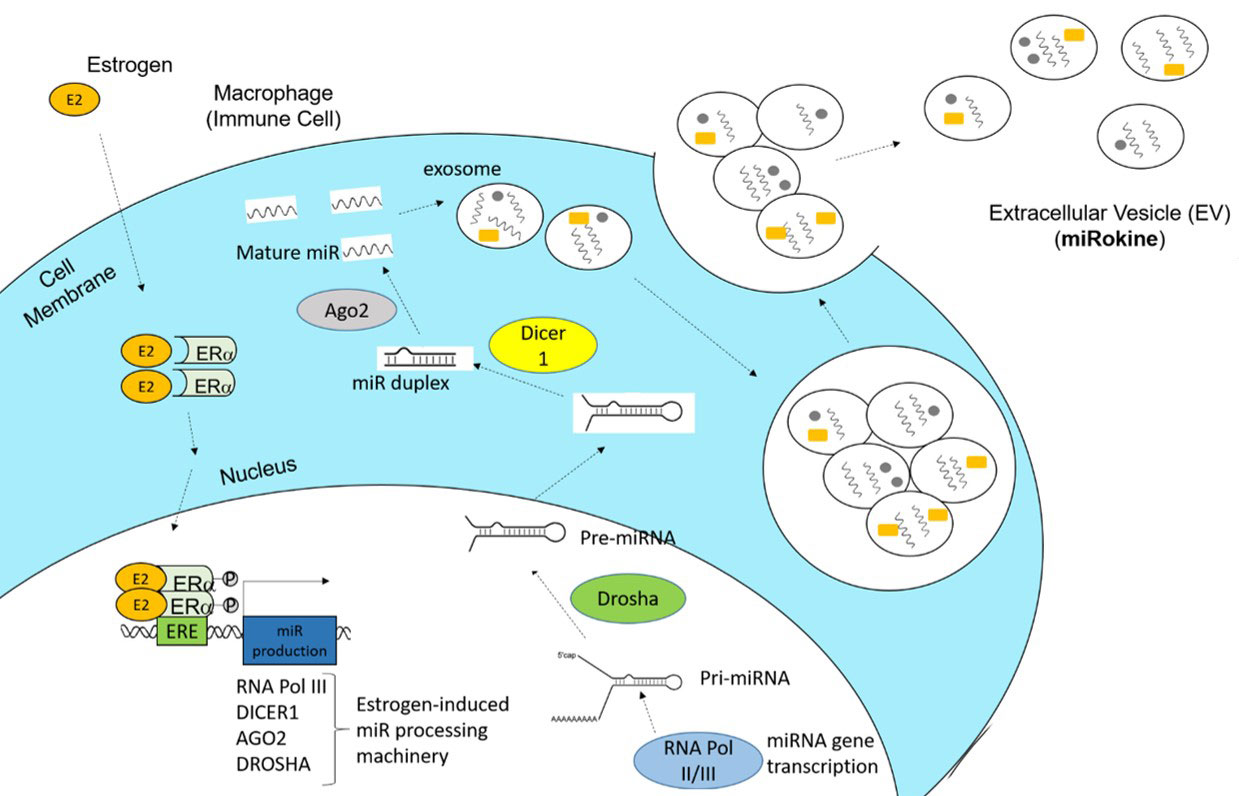
Results: Analysis of our RNAseq data by IPA revealed the predicted upregulation of estrogen-dependent breast cancer signaling by canonical pathway overlay and estrogen receptor upregulation by overlap upstream analysis. Additionally, further analysis identified upregulated pathways that included several involved in miR transcription and processing. Specifically, argonaute-2 (AGO2) had an IPA overlap p-value of 0.006, which resulted from upregulation of AGO2 expression by 1.3-fold (p < 0.01) with E2 treatment and significant downregulation of FOS, miR-127, miR-34, and miR-27. Moreover, Dicer1 (Ribonuclease III) was also stimulated 1.3-fold (p < 0.01) with E2 and an IPA overlap p-value of 0.001 was observed that resulted from miR-34, miR-196, and miR-218 suppression. Also, RNA polymerase III (p < 0.01) and Drosha RNase III (p < 0.01) were found to be signafigantly induced with estrogen treatment.
Conclusion: Our data reveal a potential mechanism of estrogen-mediated miR production and pathogenic inflammation via EV signaling in SLE. In this model, estrogen would enter an immune cell, dimerize with estrogen receptor (ER)α, translocate to the nucleus, and promote the expression of miR processing machinery, including RNA polymerase III, Dicer1, AGO2, and Drosha. The resulting miRs can then be packaged along with other proteins and secreted in EVs as extracellular signaling regulators (miRokines) that can be taken up by recipient immune cells to induce an inflammatory cascade.
To cite this abstract in AMA style:
Young N, Jablonski K, Okafor I, Schwarz E, Harb P, Henry C, Wu L, Jarjour W. RNA Sequencing of PBMCs Reveals Estrogen-Mediated Upregulation of Micro-RNA Processing Machinery [abstract]. Arthritis Rheumatol. 2019; 71 (suppl 10). https://acrabstracts.org/abstract/rna-sequencing-of-pbmcs-reveals-estrogen-mediated-upregulation-of-micro-rna-processing-machinery/. Accessed .« Back to 2019 ACR/ARP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/rna-sequencing-of-pbmcs-reveals-estrogen-mediated-upregulation-of-micro-rna-processing-machinery/