Session Information
Date: Sunday, November 7, 2021
Title: Abstracts: Epidemiology & Public Health I: Risk in Rheumatic Diseases (0988–0991)
Session Type: Abstract Session
Session Time: 4:00PM-4:15PM
Background/Purpose: The identification of lifestyle/environmental and genetic factors, influencing SLE risk introduces the potential to develop risk prediction models. We examined SLE risk prediction incorporating a lifestyle/environmental factors and a genetic risk score for SLE in predicting SLE risk among women in the prospective Nurses’ Health Studies (NHS).
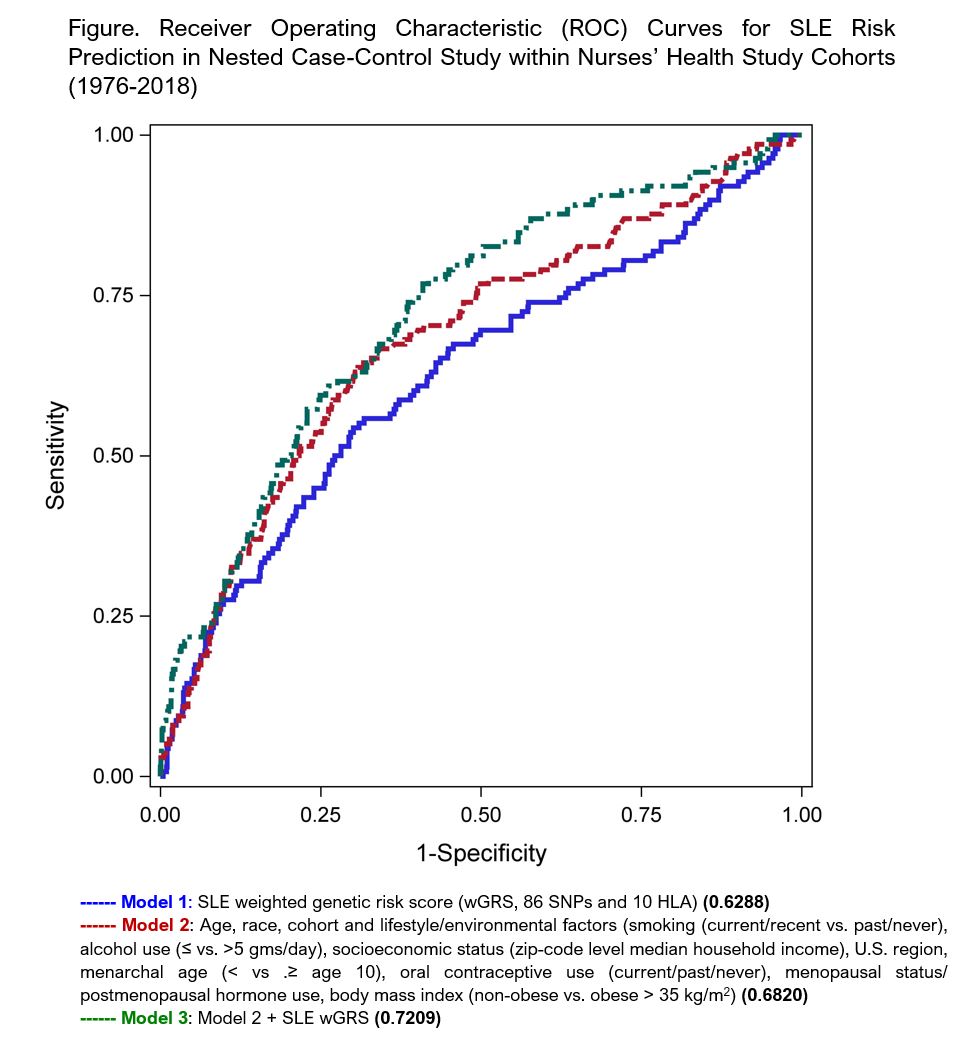
Methods: Within the prospective cohorts of female nurses, Nurses’ Health Study (NHS) (1976-2016) and NHSII (1989-2018), lifestyle, environmental, and medical data were collected at baseline and on subsequent biennial questionnaires. Incident physician diagnosed SLE was self-reported on biennial questionnaires and confirmed by medical record review to meet 1997 ACR classification criteria. Genotyping was performed on blood samples collected in 1989 (NHS) and 1997 (NHSII) from ~25% of participants. We conducted a nested case-control study in which women who had donated a blood sample and developed incident SLE were matched on age (± 4 years) and race to women who had donated a blood sample but did not develop SLE. Our previously derived and published SLE weighted genetic risk score, (wGRS, Cui J et al, Arthritis Rheum, 2019) includes 86 single nucleotide polymorphisms (SNPs) and 10 classical HLA alleles associated with SLE risk. Lifestyle/environmental variables, including smoking, alcohol use, socioeconomic status, U.S. region, menarchal age, oral contraceptive use, menopausal status/postmenopausal hormone use, body mass index, were assessed on the questionnaire prior to SLE diagnosis (or matched index date). We used multivariable logistic regression to develop sequential models of SLE risk prediction, calculating the area under the receiver operating characteristic curve (AUC). Three models were generated: 1) SLE wGRS, 2) age, race, cohort, lifestyle and environmental factors and 3) model 2 + SLE wGRS. Models were internally validated using a bootstrapped estimate of optimism of the AUC. This measure of overfitting is based on average difference between predictive ability of model using original sample, and that of models using each bootstrap sample.
Results: 138 incident cases of SLE were matched to 1136 women who did not develop SLE. Characteristics of the SLE cases and their matched controls are shown in Table. The results of our three successive prediction models are shown in Figure. The expanded model 3, including both lifestyle and genetic factors, had the best discrimination, with an AUC of 0.721 and optimism corrected AUC of 0.691.
Conclusion: A combination of lifestyle and environmental factors and weighted genetic risk score accurately classified future SLE risk with good AUC of 0.721. To our knowledge, this is the first SLE prediction model using environmental and genetic factors and might be feasibly employed in at-risk populations. The NHS cohorts include few non-White women with blood samples, and mean age at incident SLE was in early 50s given enrollment age, limiting generalizability and calling for further research in more diverse cohorts.
To cite this abstract in AMA style:
Cui J, Malspeis S, Choi M, Lu B, Sparks J, Yoshida K, Costenbader K. Risk Prediction Models for Incident Systemic Lupus Erythematosus Using Lifestyle/Environmental Risk Factors and a Genetic Risk Score [abstract]. Arthritis Rheumatol. 2021; 73 (suppl 9). https://acrabstracts.org/abstract/risk-prediction-models-for-incident-systemic-lupus-erythematosus-using-lifestyle-environmental-risk-factors-and-a-genetic-risk-score/. Accessed .« Back to ACR Convergence 2021
ACR Meeting Abstracts - https://acrabstracts.org/abstract/risk-prediction-models-for-incident-systemic-lupus-erythematosus-using-lifestyle-environmental-risk-factors-and-a-genetic-risk-score/