Session Information
Date: Monday, November 18, 2024
Title: Abstracts: Systemic Sclerosis & Related Disorders – Clinical II
Session Type: Abstract Session
Session Time: 3:00PM-4:30PM
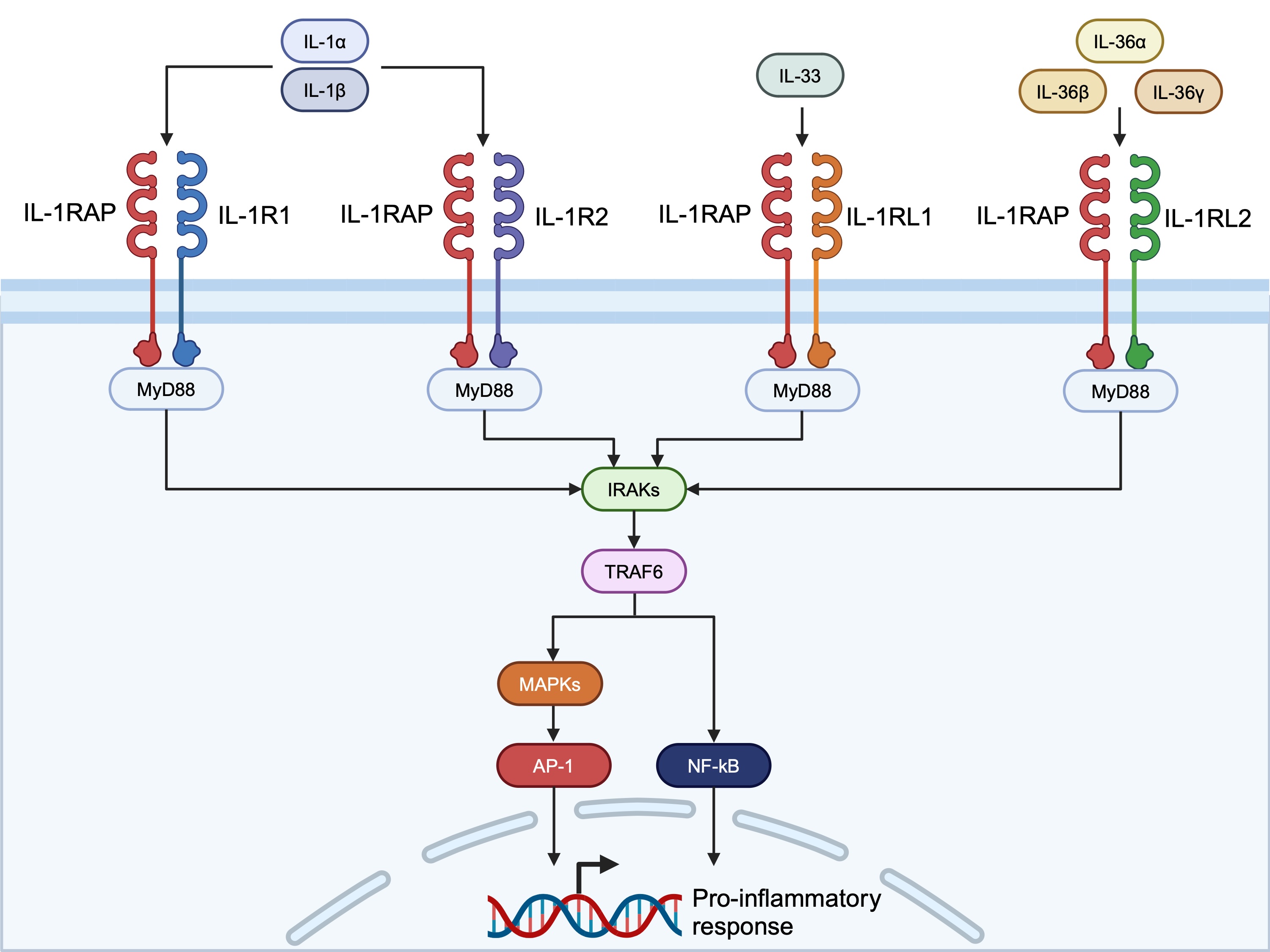
Background/Purpose: Systemic sclerosis (SSc) is an autoimmune disease characterized by systemic inflammation and fibrosis. Interleukin-1 receptor accessory protein (IL1RAP) is a co-receptor for the Interleukin-1 (IL-1) family of cytokines. It plays an essential role in the signaling of IL-1, IL-33, and IL-36, which are known to mediate inflammation and fibrosis. Recently, preclinical data indicated that the anti-IL1RAP antibody CAN10, inhibited IL-1, IL-33, and IL-36 signaling and dermal and pulmonary fibrosis. In this study, we examined the potential association of rare variants in the IL-1 family of ligands and receptors with SSc.
Methods: Exome sequences of 379 African Americans (AA) subjects with SSc and 411 AA control subjects were examined for rare variants in six ligands and five co-receptors of the IL-1 pathway (Figure 1). Replication analysis was performed in 590 AA subjects with SSc and 360 AA control subjects. The SSc subjects were enrolled at the 24 academic centers participating in the Genome Research in African American Scleroderma Patients (GRASP) consortium. Another cohort of 951 European ancestry SSc subjects and 998 EA control subjects from Spain were examined for replication. Gene-based (SKAT-O, Burden, and KBAC) testing was performed on missense, loss of start, stop gain, frameshift, or splice functional loss variants with a minor allele frequency < 0.001 in Genome Aggregation Database (gnomAD), and a Combined Annotation Dependent Depletion (CADD) score > 10 suggesting deleteriousness. Bonferroni’s multiple testing significance threshold was set at 4.5×10-3 for the discovery and 0.05 for the replication cohort.
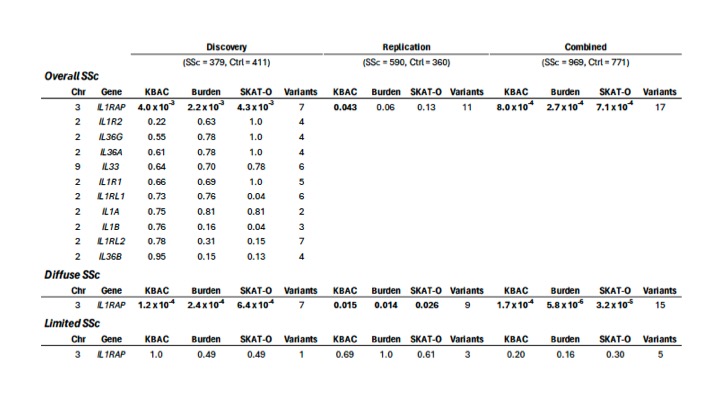
Results: All three gene-based tests identified the IL1RAP gene to be enriched for rare variants in AA subjects with SSc in the discovery and combined cohorts (Table 1). Additionally, we confirmed these findings in another large, independent, SSc cohort of European ancestry (SKAT-O p–value 3.7×10-4). Notably, diffuse cutaneous skin involvement (dcSSc) was more common in the AA subjects with SSc carrying the IL1RAP rare variants (15 of 17; 88.2%), compared to the overall AA SSc cohort (58.4%). On examining the IL1RAP association in dcSSc subset, the association was even stronger than overall SSc, whereas no association was observed in SSc patients with limited cutaneous skin involvement (Table 1). All the IL1RAP rare variants identified in SSc patients were bioinformatically predicted to be pathogenic.
Conclusion: This study, for the first time, identifies association of IL1RAP in both African and European ancestral subjects with SSc. In light of the recent preclinical data with the anti-IL1RAP antibody CAN10, prioritizing diffuse cutaneous SSc patients in clinical trials and stratifying patients based on IL1RAP variants, might pave the path for precision medicine in SSc.
IRAK; IL_1 receptor-associated kinase, MAPK; mitogen-activated protein kinase, NF-κB; nuclear factor kappa B, TRAF6; tumor necrosis factor receptor-associated factor 6.
Bold indicates statistical significance. AA; African Americans, Chr; chromosome, Ctrl; controls, KBAC; kernel-based adaptive cluster method, SKAT-O; optimal sequencing kernel association test, SSc; systemic sclerosis.
To cite this abstract in AMA style:
Kunishita Y, Kaundal U, Kerick M, Routsong R, Lack J, Shah A, Mayes M, Shriner D, Doumatey A, Bentley A, Domsic R, Medsger, Jr T, Ramos P, Silver R, Steen V, Varga J, Hsu V, Saketkoo L, Khanna D, Schiopu E, Gordon J, Criswell L, Gladue H, Derk C, Bernstein E, Bridges S, Shanmugam V, Chung L, Kafaja S, Jan R, Trojanowski M, Goldberg A, Korman B, Thomas J, Remmers E, Adeyemo A, Rotimi C, Wigley F, Boin F, Martin J, Kastner D, Gourh P. Rare Variants in the IL1RAP Gene Implicate the IL-1 Signaling Pathway in the Pathogenesis of Systemic Sclerosis in African and European Ancestries [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/rare-variants-in-the-il1rap-gene-implicate-the-il-1-signaling-pathway-in-the-pathogenesis-of-systemic-sclerosis-in-african-and-european-ancestries/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/rare-variants-in-the-il1rap-gene-implicate-the-il-1-signaling-pathway-in-the-pathogenesis-of-systemic-sclerosis-in-african-and-european-ancestries/