Session Information
Session Type: Abstract Session
Session Time: 12:15PM-12:30PM
Background/Purpose: TNFAIP3 encodes the ubiquitin editing enzyme A20, which inhibits multiple proinflammatory signaling pathways. Heterozygous germline mutations in TNFAIP3 cause the autoinflammatory disease Haploinsufficiency of A20 (HA20). The prevalence of HA20 is thought to be low, with only 200 reported cases, yet HA20 is clinically heterogeneous and has variable severity. The full spectrum of disease-causing and disease-modifying TNFAIP3 variants is unknown. Therefore, we hypothesized that HA20 prevalence is underestimated due to ascertainment bias.
Methods: To better understand HA20 prevalence and ascertain its full phenotypic spectrum, we analyzed three large population databases with whole genome sequencing data: gnomad v4 (730,947 exomes; 76,215 genomes), All of Us v7 (244,845 genomes and electronic health records (EHR) data), and UK BioBank (500,000 genomes and EHR data). We identified individuals harboring rare (MAF< 0.01%) predicted loss-of-function (pLOF) variants (frameshift, truncating, splice null) or rare predicted pathogenic missense (pPM) variants (CADD≥30; SIFT=deleterious; PolyPhen=probably damaging). Phenome-wide association studies (PheWAS) determined conditions that were significantly associated with pLOFs and pPMs. We then functionally validated the variants by performing luciferase assays to measure TNF-induced NF-kB activation, Western blot to measure basal A20 expression, and cycloheximide assays to measure protein stability. Lastly, we compared our results from these “virtual” cohorts to a “referral” cohort of patients seen at our medical center who harbor LOF (48 subjects, 17 variants) or missense (14 subjects, 9 variants) TNFAIP3 mutations.
Results: pLOF variants had a mean prevalence of 1:14,403, while pPM variants had a mean prevalence of 1:2,782, suggesting that ~100,000 to 2.9 million individuals worldwide may suffer from A20-insufficient disease. TNFAIP3 variants conferred a 164-fold risk of Behcet’s disease and a 14-fold to 138-fold risk of other HA20-associated phenotypes like stomatitis/mucositis, vasculitis, and autoimmune hemolytic anemia (PheWAS, p < 1e-5, Bonferroni). In vitro, variants reduced A20 stability, expression, and/or TNF-induced NF-kB activation. In our Pittsburgh cohort, patients exhibited clinical autoinflammation and/or autoimmunity as well as evidence of basally activated TNF-NFκB, inflammasome-IL-1β, and/or Type I interferon signaling. LOF and missense variants either increased A20 protein degradation, resulted in production of an A20 protein unable to suppress TNF-induced NFkB activation, or reduced both protein stability and function. Patients exhibited >90% response to therapies targeting A20-regulated pathways such as TNF, IL-1, and JAK inhibitors.
Conclusion: Together, these data suggest that hypomorphic TNFAIP3 variants are a massively underrecognized cause of autoinflammation with high disease burden. Increased recognition and expanded genetic testing could potentially improve health outcomes by directing these patients towards more precise immune-targeting therapies.
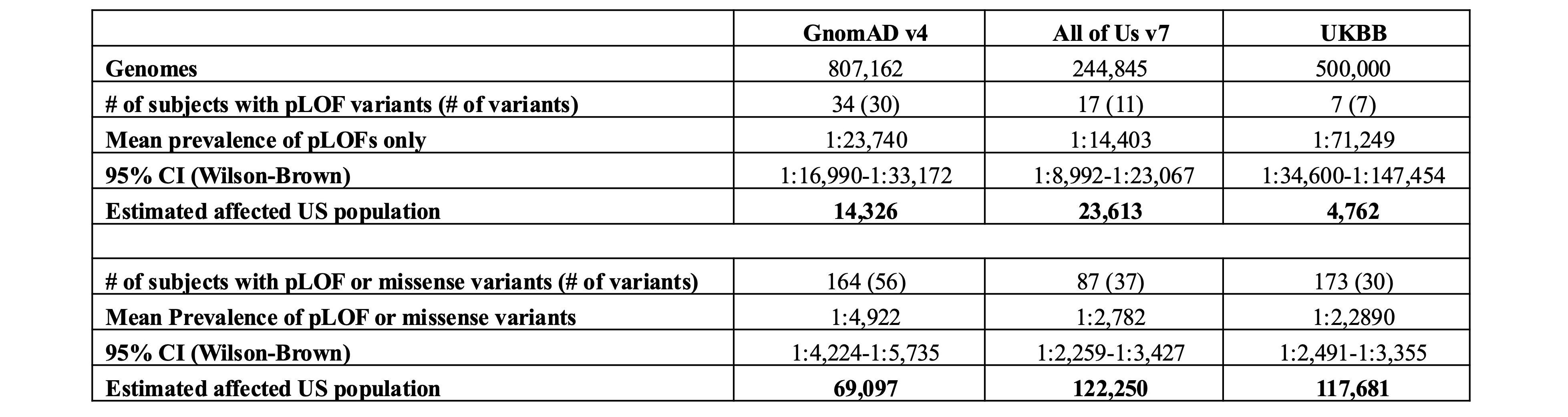 Table 1. Prevalence estimates of pLOF or missense TNFAIP3 variants in the US (AoU), UK (UKBB), and globally (gnomAD). Number of individuals harboring pLOF variants, or pPM variants (missense variants with CADD³30; SIFT=deleterious; PolyPhen=probably damaging; MAF < 0.01%). Mean prevalence of the variants and affected number of individuals were estimated based on the 2024 U.S. Census count of 340.1 million.
Table 1. Prevalence estimates of pLOF or missense TNFAIP3 variants in the US (AoU), UK (UKBB), and globally (gnomAD). Number of individuals harboring pLOF variants, or pPM variants (missense variants with CADD³30; SIFT=deleterious; PolyPhen=probably damaging; MAF < 0.01%). Mean prevalence of the variants and affected number of individuals were estimated based on the 2024 U.S. Census count of 340.1 million.
.jpg) Figure 1. Inflammatory conditions are significantly associated with pathogenic TNFAIP3 variants. Manhattan plot of conditions that are significantly associated with rare pathogenic TNFAIP3 variants in meta-PheWAS of AoU and UKBB data. Each data point represents one condition. Blue line represents uncorrected p-value and red line represents Bonferroni adjusted p-value. The table above reports odds ratio (OR) for top associated conditions compared to control cohort.
Figure 1. Inflammatory conditions are significantly associated with pathogenic TNFAIP3 variants. Manhattan plot of conditions that are significantly associated with rare pathogenic TNFAIP3 variants in meta-PheWAS of AoU and UKBB data. Each data point represents one condition. Blue line represents uncorrected p-value and red line represents Bonferroni adjusted p-value. The table above reports odds ratio (OR) for top associated conditions compared to control cohort.
.jpg) Figure 2. Hypomorphic TNFAIP3 variants can affect A20 function and/or expression. (A) Over-expressed mutant A20 proteins fail to suppress TNF-induced NF-kB activity in A20-deficient HEK293 cells transfected with the luciferase reporter plasmid (*p < 0.05, ratio paired t-test); WT = wild-type, EV = empty vector. (B) Basal A20 expression (Western blot) of A20 variants, expression normalized to actin.
Figure 2. Hypomorphic TNFAIP3 variants can affect A20 function and/or expression. (A) Over-expressed mutant A20 proteins fail to suppress TNF-induced NF-kB activity in A20-deficient HEK293 cells transfected with the luciferase reporter plasmid (*p < 0.05, ratio paired t-test); WT = wild-type, EV = empty vector. (B) Basal A20 expression (Western blot) of A20 variants, expression normalized to actin.
To cite this abstract in AMA style:
Lee D, Karri U, Luo Y, Cetin Gedik K, Carpio Tumba M, Chhibbar P, Roy P, Falduto G, Das J, Schwartz D. Rare TNFAIP3 Hypomorphic Variants are a Massively Underestimated Driver of Human Autoinflammatory Disease [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/rare-tnfaip3-hypomorphic-variants-are-a-massively-underestimated-driver-of-human-autoinflammatory-disease/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/rare-tnfaip3-hypomorphic-variants-are-a-massively-underestimated-driver-of-human-autoinflammatory-disease/
