Session Information
Date: Sunday, October 26, 2025
Title: (0098–0114) Spondyloarthritis Including Psoriatic Arthritis – Basic Science Poster
Session Type: Poster Session A
Session Time: 10:30AM-12:30PM
Background/Purpose: While numerous proteins have been linked to psoriatic arthritis (PsA), the causal nature of these associations remains unconfirmed. This study aims to employ a Mendelian randomization (MR) approach to evaluate whether circulating plasma protein levels, derived from extensive international databases, are causally associated with PsA.
Methods: The study analyzed 1,553 PsA patients and 147,221 controls, all of whom had undergone genome-wide association scans, as identified from the FinnGen consortium. Large-scale plasma protein quantitative trait locus (pQTL) data were sourced from the UK Biobank Proteomics Project (UKB-PPP) (2,923 unique proteins), the INTERVAL study (2,995 unique proteins), and the Icelandic study (Ferkingsatad et al. 2021) with (4,719 proteins). Potential protein candidates were identified based on SNP associations with proteins (p < 5×10^-8), LD clumping to identify independent pQTLs for each protein (r^2 < 0.001), and defining SNPs when the leading SNP was located within 500kb of the transcription site of the protein-coding gene. A two-sample MR was then performed, using the Wald ratio for genes with one SNP and inverse variance weighting (IVW) for genes with more than one SNP. Appropriate MR sensitivity and pleiotropy analyses were conducted.
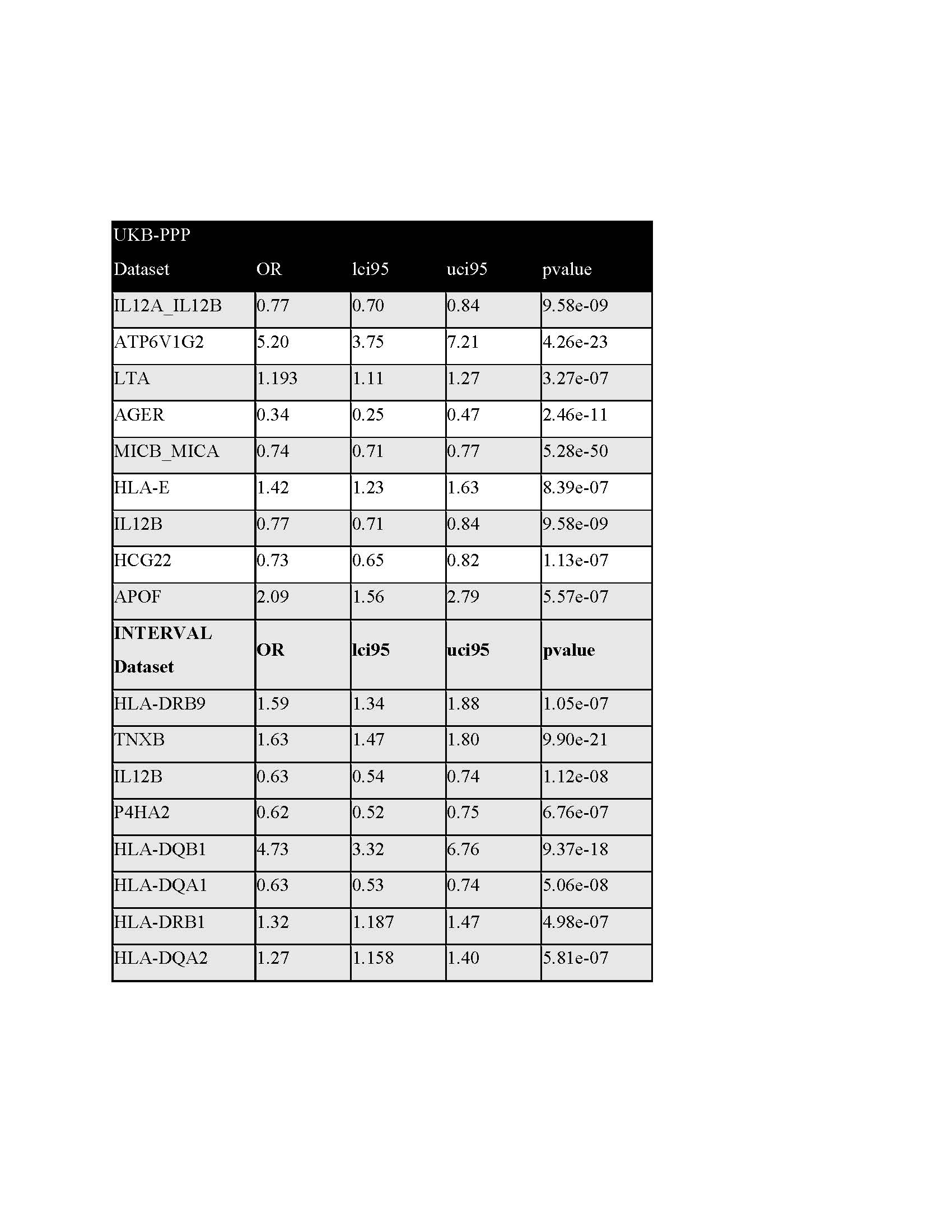
Results: A total of 19 unique proteins were identified as being associated with the risk of PsA. UKB-PPP dataset revealed four proteins linked to an increased risk of PsA (ATP6V1G2, LTA, HLA-E, APOF) and five proteins associated with a decreased risk (IL12A_IL12B, AGER, MICB_MICA, IL12B, HCG22) (Table 1). The INTERVAL dataset identified five proteins correlated with an elevated risk of PsA (HLA-DRB9, TNXB, HLA-DQB1, HLA-DRB1, HLA-DQA2) and three proteins linked to a reduced risk (IL12B, P4HA2, HLA-DQA1) (Table 1). From Ferkingsatad et al. 2021 dataset, MR identified five proteins significantly associated with PsA (C2,TNXB,HSPA1A and APOF increased PsA risk, while AGER decreased the risk, Table 2).A protein-protein interaction network was constructed using the STRING database, with a required interaction score of >0.4, illustrating the connections among these proteins. Notably, many of these proteins were found within the drug repurposing hub, therapeutic target database, and druggable genome compendium.
Conclusion: This proteome-wide MR study identified 19 unique proteins associated with PsA risk, offering novel insights into the disease’s pathogenesis. These prioritized proteins also present potential druggable targets, warranting further investigation to explore novel therapeutic opportunities. More in depth studies are needed to assess the role and utility of these proteins.
To cite this abstract in AMA style:
Li Q, Rahman P. Proteome-wide Mendelian Randomization Identifies Therapeutic Targets in Psoriatic Arthritis [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/proteome-wide-mendelian-randomization-identifies-therapeutic-targets-in-psoriatic-arthritis/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/proteome-wide-mendelian-randomization-identifies-therapeutic-targets-in-psoriatic-arthritis/

.jpg)
.jpg)