Session Information
Date: Sunday, November 10, 2019
Title: RA – Diagnosis, Manifestations, & Outcomes Poster I: Risk Factors, Predictors, & Prognosis
Session Type: Poster Session (Sunday)
Session Time: 9:00AM-11:00AM
Background/Purpose: Determining the prognosis of rheumatoid arthritis (RA) could guide more precise treatment selection, optimizing outcomes while minimizing unnecessary therapeutic exposures. It is important that potential biomarkers have widespread availability and provide meaningful input in clinical decision making. While CRP and ESR have been shown to be moderately associated with disease activity and severity, there continues to be uncertainty regarding the utility of these as they may be elevated in other conditions, or be normal even during severe clinical inflammation. While RF and anti-CCP positivity are considered to be associated with more treatment refractory RA, those who are autoantibody negative may experience a negative disease trajectory as well due to lack of recognition of disease and introduced delays in treatment initiation. Our objective was to employ principal component analysis to investigate if biomarkers could determine sub-groups of disease severity.
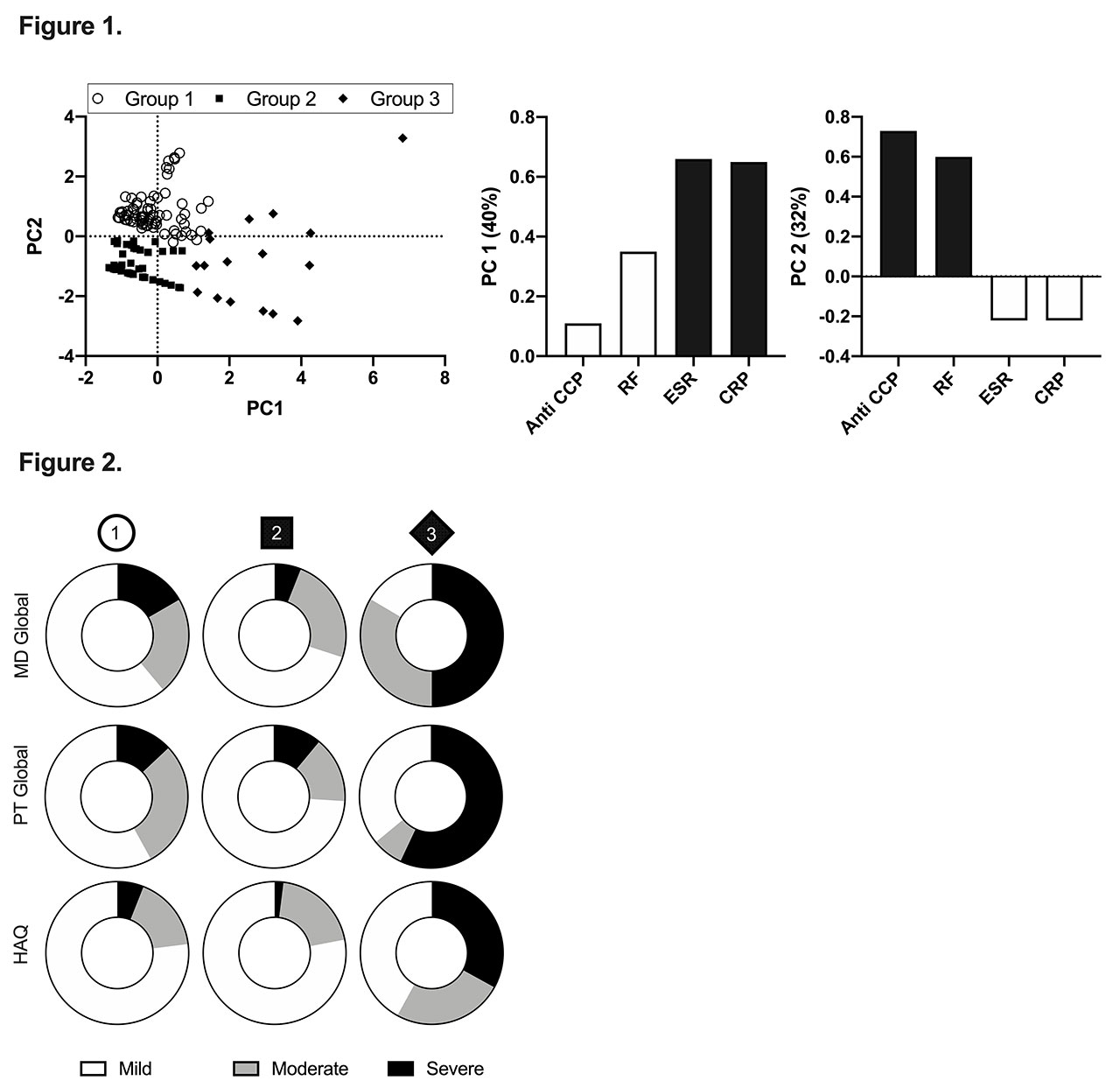
Methods: RA patients were recruited from a single centre to a cross sectional study initiated to investigate influences on disease trajectory (n=145). ESR, CRP, RF, and anti-CCP collected at enrolment to the study were used in the principle component analysis (PCA) to create 2 components, which were subsequently used in k-means clustering (K=3) to demarcate the patient groups. Function (Health Assessment Questionnaire), Physician Global Assessments (MD Global), and Patient Global Assessments (Pt Global) were divided into 3 tertiles of disease activity (mild, moderate, and severe) and used as metrics of disease severity to describe the patient groups identified by PCA and K-means.
Results: Based on correlation analysis, ESR and CRP had a moderate positive relationship with each other (r=0.67), but no correlation with HAQ, Pt Global, or MD Global. Furthermore, there was no relationship between individual serological markers and disease activity as reported by HAQ, Pt Global, or MD Global (r< 0.25 for all). PCA revealed the presence of three visual groupings of data points (PC1, PC2 and PC3), that were then confirmed with K-means clustering (Figure 1). ESR and CRP weighed most heavily in PC1, while PC2 was predominantly influenced by anti-CCP and RF. Disease severity assessments illustrate that PC3 had a higher percentage of severe disease and PC2 showed a higher percentage of mild disease (Figure 2).
Conclusion: Sub-populations with unique serological biomarker combinations that predict disease severity are identifiable. The sub-group that was not influenced as strongly by serology was the group identified to have the largest proportion of patients with mild disease. Furthermore, the group with the highest proportion of patients with severe disease was most heavily influenced by ESR and CRP, and the group with the next highest percentage of severe disease was influenced predominantly by anti-CCP and RF. This suggests 2 unique populations with higher serological values and various weighted combinations of ESR and CRP, or anti-CCP and RF, seem to display RA with a more severe phenotype. Further studies will need to be undertaken to identify factors that make these RA groups unique.
To cite this abstract in AMA style:
Van Dyk J, Heard B, Barnabe C. Principal Component Analysis Identifies Unique Sub-Populations in Rheumatoid Arthritis Using a Combination of Serological Biomarkers: A Cross Sectional Study [abstract]. Arthritis Rheumatol. 2019; 71 (suppl 10). https://acrabstracts.org/abstract/principal-component-analysis-identifies-unique-sub-populations-in-rheumatoid-arthritis-using-a-combination-of-serological-biomarkers-a-cross-sectional-study/. Accessed .« Back to 2019 ACR/ARP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/principal-component-analysis-identifies-unique-sub-populations-in-rheumatoid-arthritis-using-a-combination-of-serological-biomarkers-a-cross-sectional-study/