Session Information
Date: Tuesday, November 15, 2016
Title: Metabolic and Crystal Arthropathies - Poster II: Epidemiology and Mechanisms of Disease
Session Type: ACR Poster Session C
Session Time: 9:00AM-11:00AM
Background/Purpose
: Accurate case-definition is important for epidemiological studies of gout. However, in multipurpose cohort studies frequently used for genome wide association studies, limited information is usually available for gout case-definition. Many different combinations of available data have been used to identify gout cases in large genetic epidemiological studies. The aim of this study was to determine the most precise case-definition of gout from the limited items available in multipurpose cohorts for use in for population-based genetic studies.
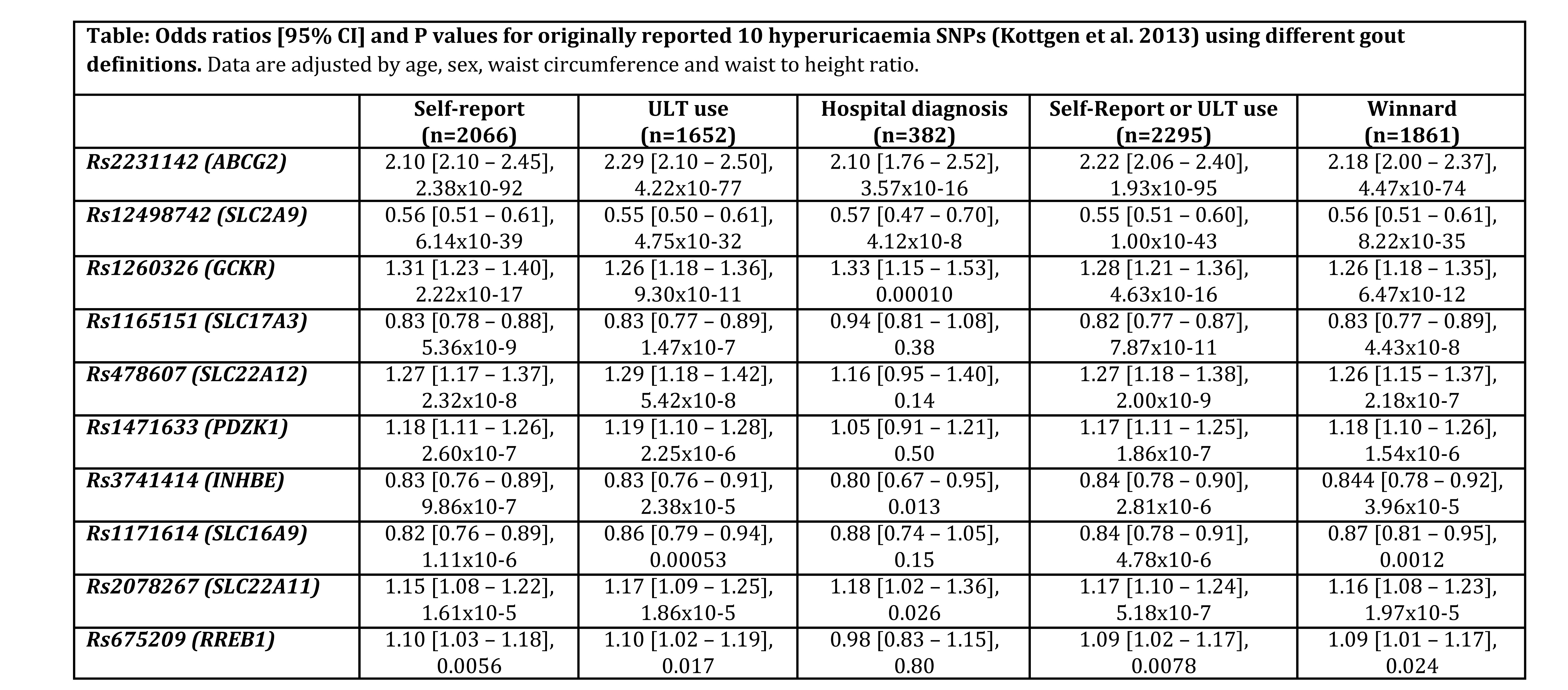
Methods: Data from the UK Biobank were analysed. Data (including genome-wide genotypes) were available for 105,421 European participants aged 40 to 69 years without kidney disease. Gout definitions and combinations of these definitions were identified from previous epidemiological studies. Self-report of gout was defined by reporting of gout diagnosis by the participant at the time of study interview. Hospital diagnosis of gout was defined by either primary or secondary hospital discharge coding for gout (ICD10 M10 including subcodes). Urate lowering therapy (ULT) use was defined by self-report of allopurinol, febuxostat, or sulphinpyrazone use, without leukemia or lymphoma (ICD10 C81-C96). Winnard-defined gout was defined by hospital diagnosis or gout specific medication (ULT or colchicine)1. Logistic regression was performed genome-wide using plink2 adjusted for age, sex, waist circumference, and waist circumference to height ratio.
Results : There were 2066 (2.0%) gout cases defined by self-report of gout, 1652 (1.6%) defined by ULT use, 382 (0.4%) defined by hospital diagnosis, 2295 (2.2%) defined by self-report of gout or ULT use, and 1861 (1.8%) gout cases defined by Winnard-definition. For the originally reported 10 hyperuricaemia SNPs2, association with gout at genome-wide significance (P<5×10-8) was observed for 5 SNPs (ABCG2, SLC2A9, GCKR, SLC17A3 and SLC22A12) using the definition of self-report of gout and the definition of self-report of gout or ULT use, 4 SNPs (ABCG2, SLC2A9, GCKR, and SLC17A3) using the Winnard definition, 3 SNPs (ABCG2, SLC2A9, and GCKR) using the ULT use definition, and 2 SNPs (ABCG2 and SLC2A9) using the hospital diagnosis definition (Table). The definitions of self-report of gout or ULT use, and self-report of gout alone had greatest precision for all analyses.
Conclusion: Of existing definitions that use routinely collected data, case definitions that include self-report of gout have the greatest precision for detecting genome wide significant associations in epidemiological studies of gout. References: 1. Winnard et al Rheumatology 2012, 2. Kottgen et al Nature Genetics 2013.
To cite this abstract in AMA style:
Cadzow M, Merriman TR, Dalbeth N. Precision of Gout Definitions for Population-Based Genetic Studies: Analysis of the UK Biobank [abstract]. Arthritis Rheumatol. 2016; 68 (suppl 10). https://acrabstracts.org/abstract/precision-of-gout-definitions-for-population-based-genetic-studies-analysis-of-the-uk-biobank/. Accessed .« Back to 2016 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/precision-of-gout-definitions-for-population-based-genetic-studies-analysis-of-the-uk-biobank/