Session Information
Session Type: Poster Session (Tuesday)
Session Time: 9:00AM-11:00AM
Background/Purpose: Patients with SLE have increased risk of cardiovascular events and a higher prevalence of metabolic conditions compared to the general population. Inflammation is a strong component of the pathogenesis of SLE, cardiometabolic disorders, and other autoimmune diseases; thus, we examined the hypotheses that genetic predisposition to SLE may increase the risk for: 1) cardiometabolic outcomes and 2) other autoimmune diseases.
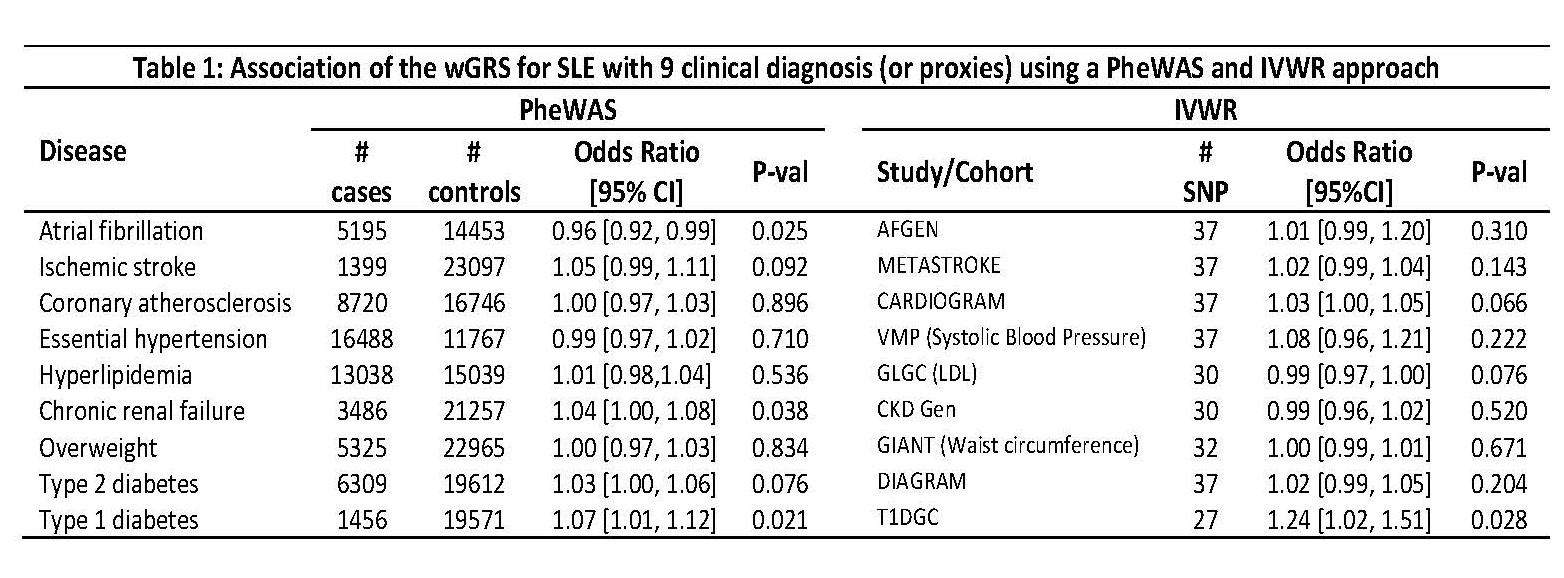
Methods: To test these hypotheses, we selected 47 independent SNPs (R2< 0.10) in 36 loci with significant associations with SLE (P< 5×10-8) in the largest genetic study performed in Caucasians. In the Vanderbilt DNA biobank (BioVU), forty-one SNPs had a call rate ≥ 95% and were used to construct a weighted genetic risk score (wGRS) for SLE to: 1) test its association with 9 cardiometabolic disorders (Table 1) and 2) examine the relationship between the wGRS and all phenotypes present in 29,892 Caucasian patients in a global phenome wide association study (PheWAS) adjusting for median age in the electronic health record, sex, and 5 principal components. A P-value ≤ 0.006 was considered significant for the 9 selected cardiometabolic diseases, and a false discovery rate (FDR) < 0.1 was used in the global PheWAS. In addition, a set of independent SLE-associated SNPs (R2 < 0.05) was used to perform a two-sample inverse-variance weighted regression (IVWR) meta-analysis using publicly available summary statistics for the 9 cardiometabolic phenotypes of interest (or proxies). Egger regression was performed to test for horizontal pleiotropy with phenotypes in which the IVWR analysis was significant.
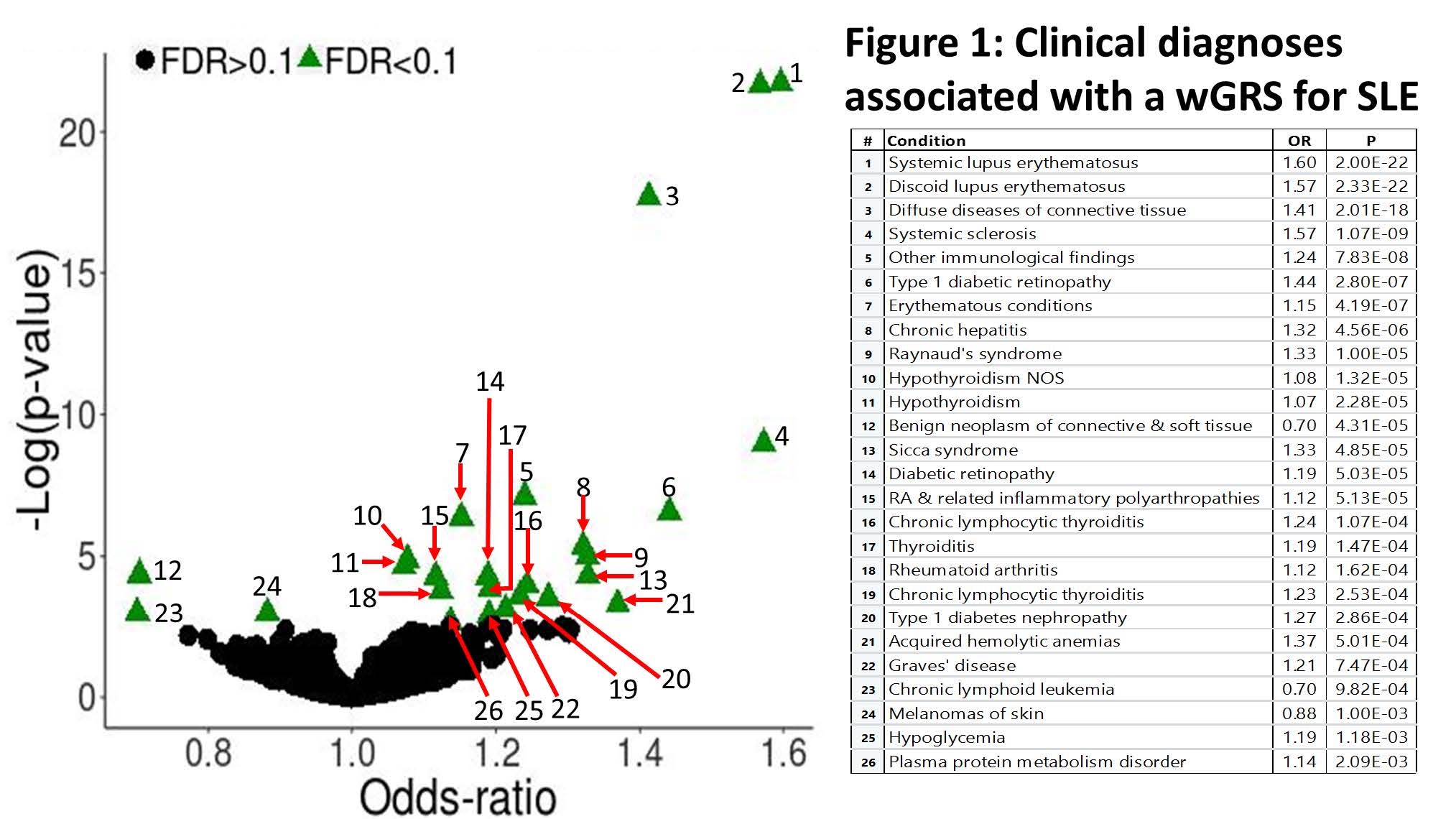
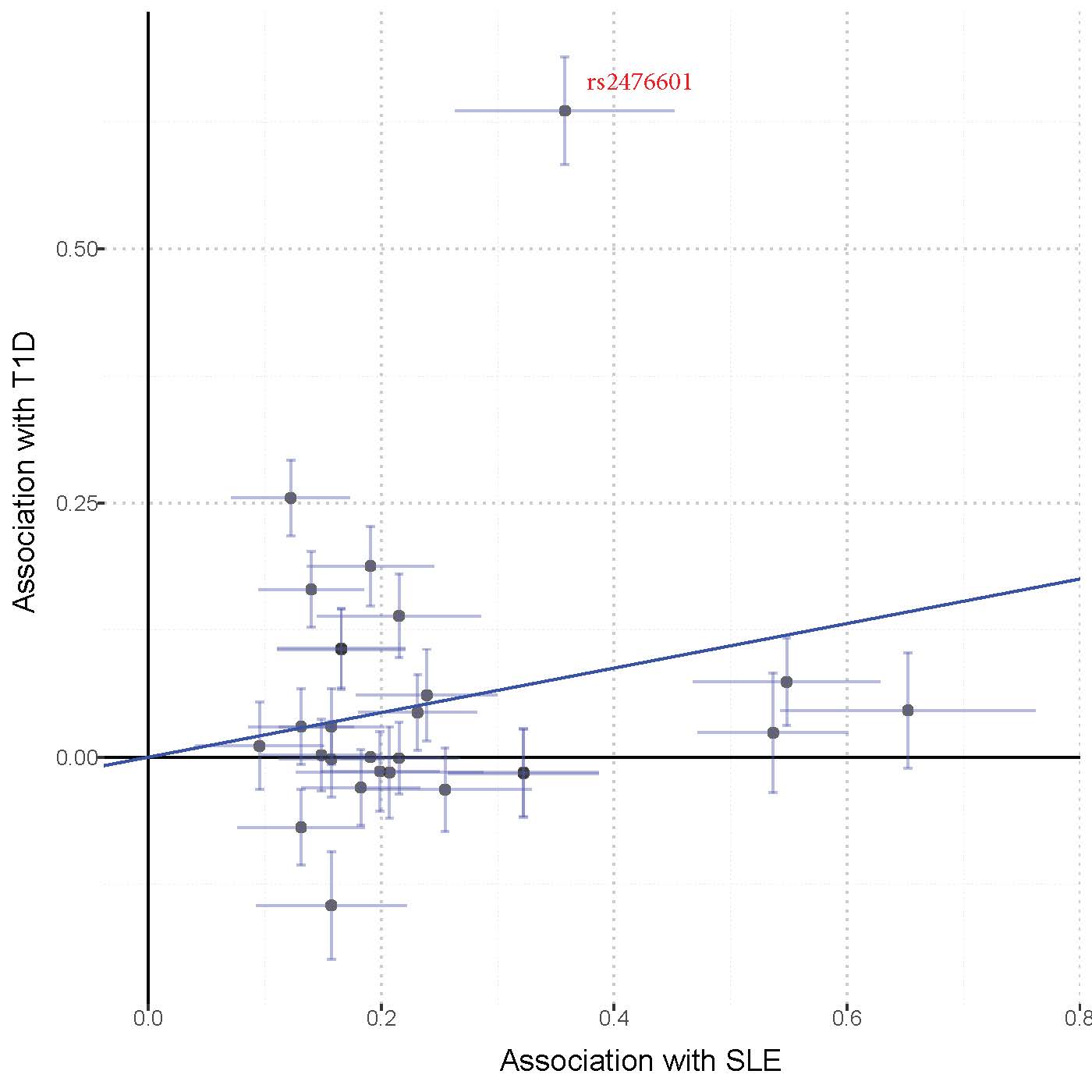
Results: There was no significant association between the SLE wGRS and the 9 selected cardiometabolic phenotypes (all P values >0.006, Table 1). In the global PheWAS the wGRS was associated with several autoimmune phenotypes including SLE (OR 1.6, P=2×10-22), systemic sclerosis, hypothyroidism, Grave’s disease, rheumatoid arthritis, and several T1D phenotypes (Fig. 1). The IVWR analyses comparing the genetics of SLE with that of the 9 selected phenotypes was significant for T1D (P=0.028, Table 1; Fig. 2). In the Egger analysis, the intercept term of the regression was not significant (P=0.49) suggesting no horizontal pleiotropy between SLE and T1D. Exclusion of the SLE-variant rs2476601, a SNP in PTPN22, in the T1D analysis (Fig. 2) attenuated the results of the IWVR analysis (P=0.06).
Conclusion: We found evidence suggesting pleiotropy between genetic predisposition for SLE and several autoimmune disorders as well as diabetic retinopathy and nephropathy. In the IVWR analysis, genetic predisposition to SLE was associated with increased risk of T1D, in part through a common missense variant in PTPN22 known to be associated with several autoimmune phenotypes.
To cite this abstract in AMA style:
Kawai V, Shi M, Feng Q, Chung C, Liu G, Cox N, Roden D, Stein C, Mosley J. Pleiotropy of Systemic Lupus Erythematosus (SLE) Risk Alleles: Association with Increased Risk for Type 1 Diabetes (T1D) Complications Through a PTPN22 Polymorphism [abstract]. Arthritis Rheumatol. 2019; 71 (suppl 10). https://acrabstracts.org/abstract/pleiotropy-of-systemic-lupus-erythematosus-sle-risk-alleles-association-with-increased-risk-for-type-1-diabetes-t1d-complications-through-a-ptpn22-polymorphism/. Accessed .« Back to 2019 ACR/ARP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/pleiotropy-of-systemic-lupus-erythematosus-sle-risk-alleles-association-with-increased-risk-for-type-1-diabetes-t1d-complications-through-a-ptpn22-polymorphism/