Session Information
Session Type: Poster Session A
Session Time: 10:30AM-12:30PM
Background/Purpose: Antiphospholipid syndrome (APS) is an autoimmune disease with thromboembolic and obstetric morbidity arising via a model of immunothrombosis. Patients may present with thrombotic (tAPS), obstetric (oAPS), or catastrophic/microvascular (C/MAPS) disease, while many have circulating antiphospholipid antibodies (aPL) without clinical APS. To understand peripheral biologic mechanisms underlying different APS phenotypes, we profiled plasma proteome of persistently aPL-positive patients without other systemic autoimmune diseases, and present expanded analyses together with a validation cohort.
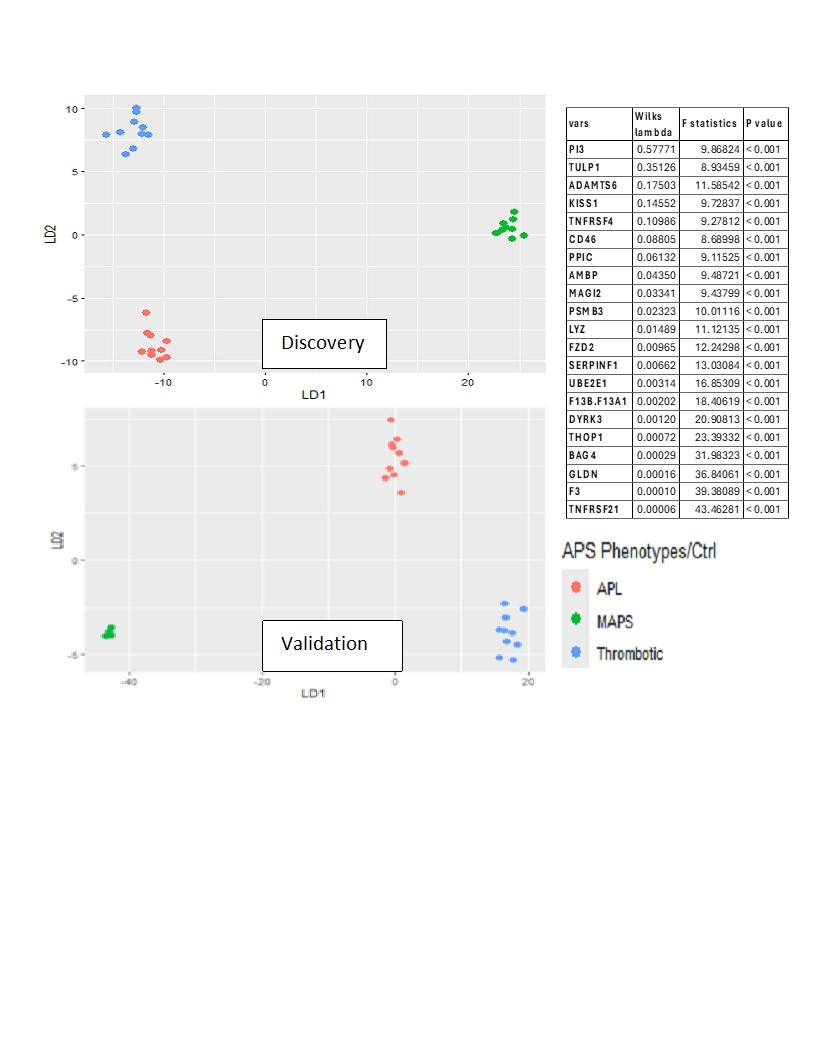
Methods: Plasma samples were obtained from APS ACTION Registry. Discovery cohort (DC): 10 each of tAPS, oAPS, C/MAPS, and aPL-only; 10 healthy controls; 6,398 unique proteins assayed using SomaScan. Differential proteins were determined by ANOVA and t-tests in log-normalized data (p-values< 0.05, false discovery rate q< 0.1) and functional enrichment analysis performed. Validation cohort: 10 tAPS, 10 oAPS, 4 C/MAPS, and 10 aPL-only; 1500 proteins derived from the DC assayed by SomaScan. Proteins likely to discriminate aPL, TAPS, and C/MAPS groups were derived from DC with stepwise feature selection using Wilks lambda criterion. The clustering accuracy of the identified protein set was then tested using linear discriminant analysis (LDA).
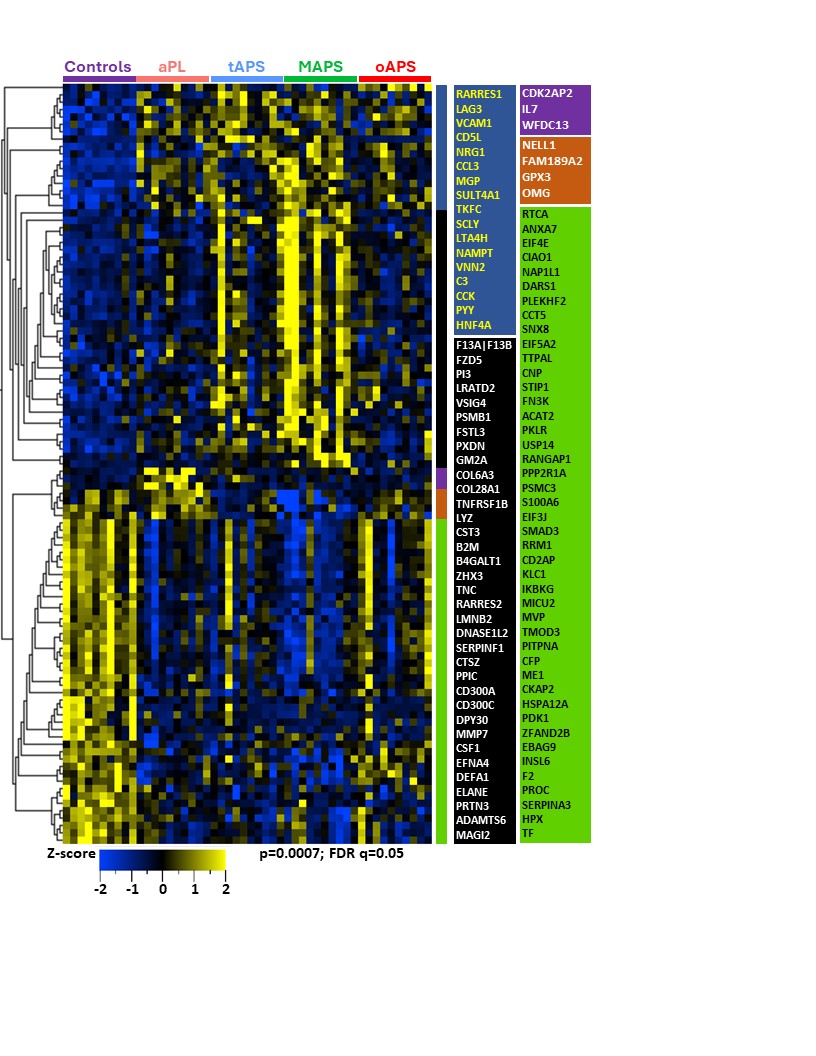
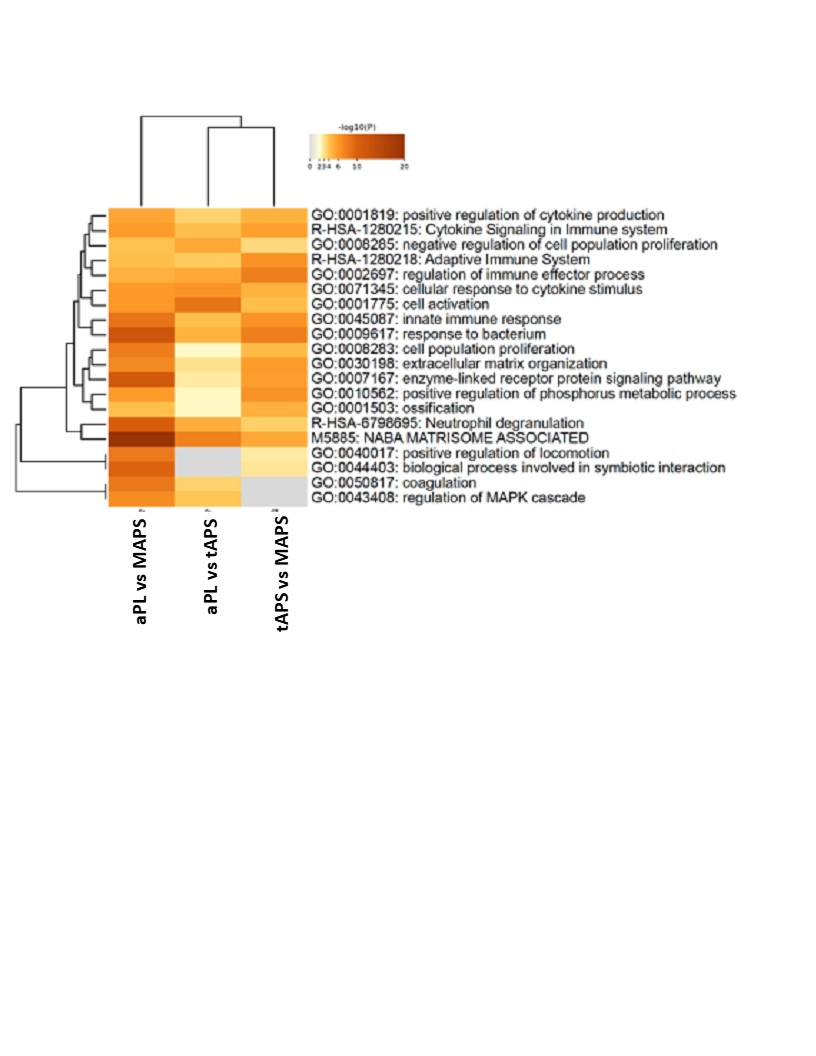
Results: The median age was 48 years, 30% were men, and 70% were triple aPL-positive. A set of proteins clustered individuals with four APS clinical phenotypes and controls (Figure 1; p< 0.0007; q< 0.05). Differential proteins belonged to highly enriched pathways including coagulation, complement, innate and adaptive immunity. Strikingly, the aPL-only cohort shared this thromboinflammatory signature with other groups, while oAPS appeared distinct from tAPS and C/MAPS. Pathway analysis of protein sets identified from pairwise comparisons (aPL vs tAPS, aPL vs C/MAPS, tAPS vs C/MAPS) revealed progressive enrichment and increasing statistical significance of extracellular matrix (ECM) signaling, coagulation, neutrophil degranulation, innate and adaptive immune responses, and cellular and cytoskeletal activation pathways, suggesting a model of increasing thromboinflammation, including tissue inflammation, in evolution from aPL to C/MAPS (Figure 2). Unbiased clustering of the discovery cohort by plasma proteomics revealed distinct clustering by APS subtype (Figure 3). TNFRSF21, tissue factor, BAG4, factor 13, lysozyme, pigment epithelium-derived factor, and TNFRSF4 were top hits among 21 proteins featured in this analysis. Importantly, these proteins successfully clustered the validation cohort by APS subtype.
Conclusion: A mere presence of aPL confers a distinct thromboinflammatory signature characterized by coagulation, complement, innate and adaptive immune response pathways shared by all APS subtypes with an increasing frequency of abnormalities in C/MAPS. Progressive thromboinflammation together with involvement of the ECM, intracellular immune cell signaling, cytoskeletal organization and vesicular trafficking underlies evolution of APS from aPL to C/MAPS.
To cite this abstract in AMA style:
Pine A, Pengo V, Sciascia S, Kello N, Lopez Pedrera R, Belmont H, Branch D, Andreoli L, Petri M, Cervera R, Knight J, Meroni P, Cohen H, Willis R, Bertolaccini M, Lee A, Erkan D, Sharda A. Plasma Proteomics Analysis Identifies Thromboinflammatory Signature Associated with Clinical Antiphospholipid Syndrome: Results from Antiphospholipid Syndrome Alliance for Clinical Trials and International Networking (APS ACTION) Registry [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/plasma-proteomics-analysis-identifies-thromboinflammatory-signature-associated-with-clinical-antiphospholipid-syndrome-results-from-antiphospholipid-syndrome-alliance-for-clinical-trials-and-internat/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/plasma-proteomics-analysis-identifies-thromboinflammatory-signature-associated-with-clinical-antiphospholipid-syndrome-results-from-antiphospholipid-syndrome-alliance-for-clinical-trials-and-internat/