Session Information
Date: Monday, November 14, 2022
Title: Abstracts: SLE – Diagnosis, Manifestations, and Outcomes III: Genetic Factors
Session Type: Abstract Session
Session Time: 3:00PM-4:30PM
Background/Purpose: Prior studies show an independent association between greater physical activity and lower inflammatory markers among adults in the general population, but the impact of physical activity on systemic inflammation in people with systemic lupus erythematosus (SLE) is unknown. We aimed to determine whether physical activity is associated with cell-specific differential expression of inflammatory genes in SLE.
Methods: Data derive from the California Lupus Epidemiology Study (CLUES), a prospective observational cohort of adults meeting ACR criteria for SLE. Physical activity was assessed by self report: those who endorsed being “seldom active” were classified as “inactive” while those who reported moderate or vigorous physical activity for ≥30 minutes on ≥3 days/week were classified as “active”. Peripheral blood obtained at study visits was sorted utilizing magnetic beads into four cell types: CD14+ monocytes, CD19+ B cells, CD4+ T cells, and NK cells. RNA extraction from sorted cells, library preparation, sequencing, and gene expression data processing were performed using established methods. Genes differentially expressed in the sedentary versus active groups were identified using DESeq2 with the Independent Hypothesis Weighting (IHW) multiple testing procedure. We adjusted for sex, age, race, and ethnicity in the final multivariable linear model. To identify biological pathways most affected by physical activity, Ingenuity Pathway Analysis (IPA) Upstream Regulator Analyses were employed on differentially expressed genes ranked by log2 fold change.
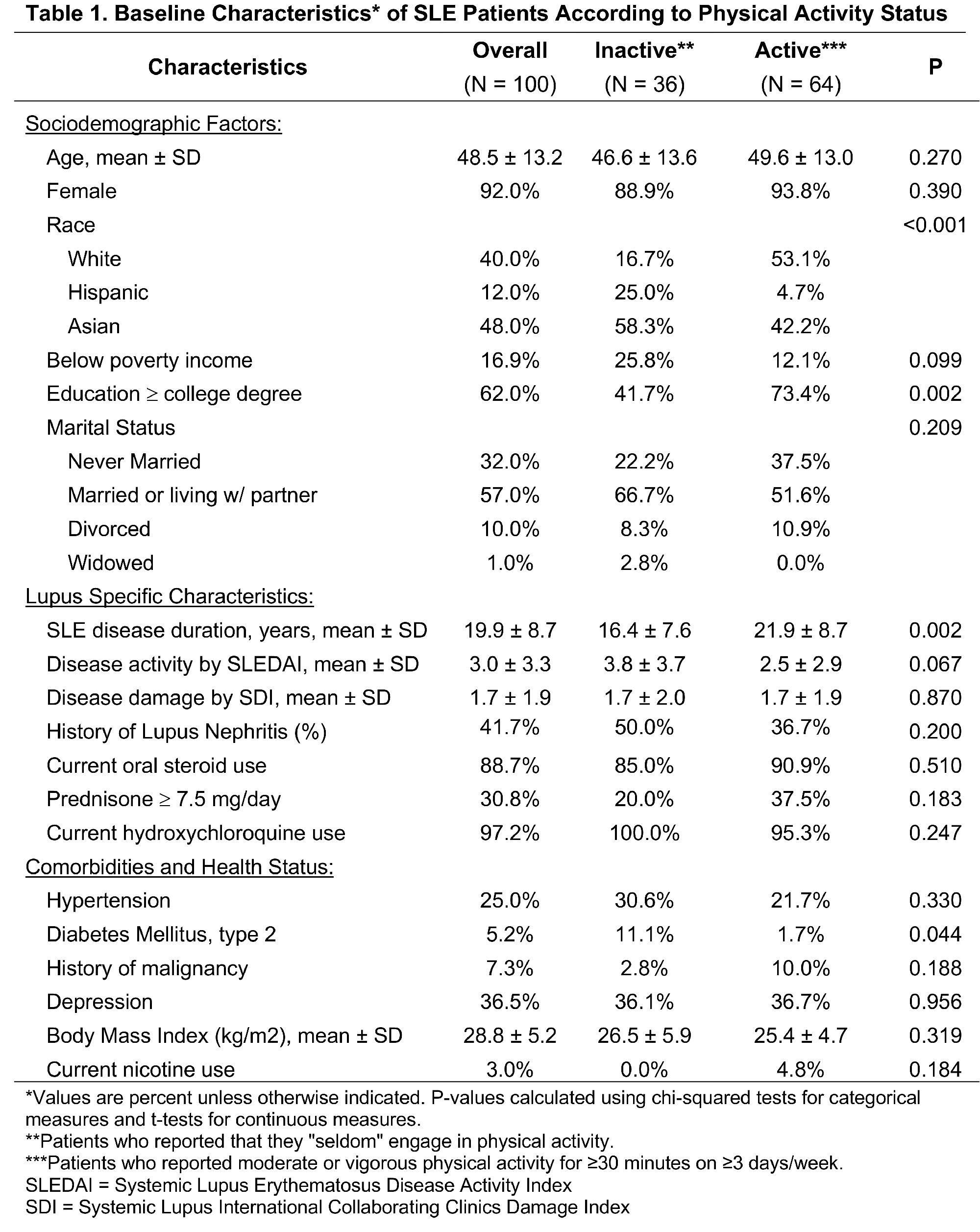
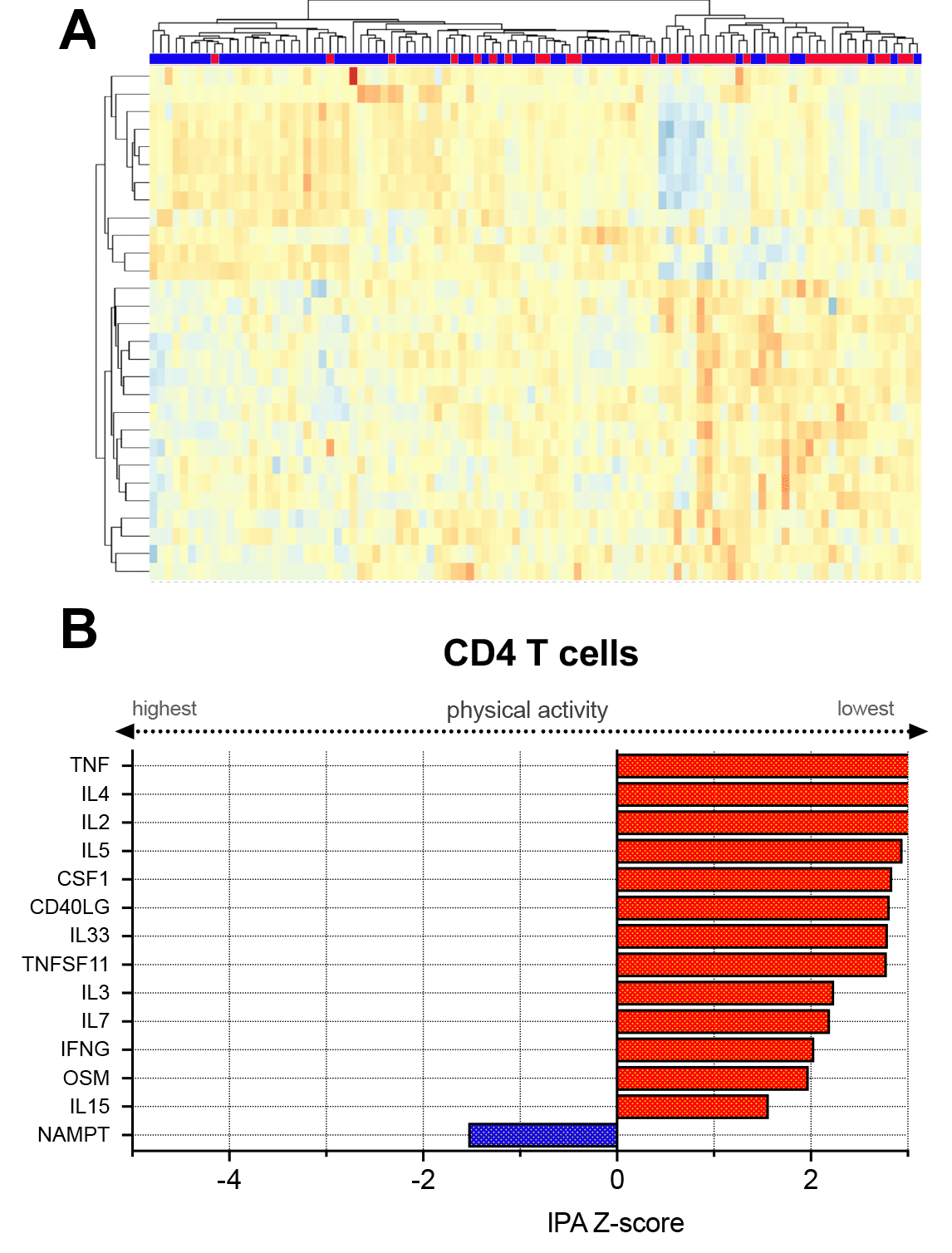
Results: 100 patients had complete clinical, physical activity, and transcriptomic data available for analysis. Mean age was 48.5 years (SD 13.2), and 92% were female. Race/ethnicity was 40% white, 48% Asian, and 12% Hispanic (Table 1). 36% of the cohort reported that they were seldom active while the remaining 65% engaged in regular moderate or vigorous physical activity. Among CD4+ T cells, there were 1,022 genes differentially expressed (DE) between the inactive versus physically active groups at an adjusted P value < 0.1 (Figure 1a). Within NK T cells there were 156 DE genes, whereas there were no DE genes in the CD19+ and CD14+ cell types by physical activity status. Assessment of upstream cytokine activation states in CD4+ T cells demonstrated activation of several cytokines involved in immune signaling—including TNF, CD40 ligand (CD40L), IL-4, IL-2, and interferon-g (IFNG)—among sedentary patients compared to those who were more physically active (Figure 1B). Similarly, within NK cells, sedentary patients had upstream cytokine activation of IL-33, TNF, IL1-b, IL-17, and IL-23 compared to the physically active patients.
Conclusion: Among a racially and ethnically diverse SLE cohort, participants who were sedentary had upregulation of genes involved in both innate and adaptive immune signaling in CD4+ and NK T cells compared to those who engaged in regular physical activity. These findings provide mechanistic evidence that physical inactivity may augment T-cell-mediated systemic inflammation in SLE.
a) Heatmap demonstrating unsupervised clustering of top differentially expressed genes in CD4+ T cells based on physical activity group. Participants are indicated in the top row: red represents the sedentary patients and light blue represents physically active patients. In the heatmap, genes upregulated are represented in orange while genes downregulated are represented in light blue.
b) Predicted activation state of upstream cytokines in CD4+ T cells among sedentary versus physically active patients. Cytokines with an IPA |Z| > 2 are plotted. Upregulated cytokines have positive IPA Z-scores and are represented in red. Downregulated cytokines have negative Z-scores and are represented in dark blue.
To cite this abstract in AMA style:
Patterson S, Cruz Gonzalez S, Tsitsiklis A, Lanata C, Maliskova L, Ye C, Cutts Z, Yazdany J, Dall'Era M, Criswell L, Langelier C, Katz P, Sirota M. Physical Inactivity Is Associated with Increased Expression of T Cell Inflammatory Genes in Systemic Lupus [abstract]. Arthritis Rheumatol. 2022; 74 (suppl 9). https://acrabstracts.org/abstract/physical-inactivity-is-associated-with-increased-expression-of-t-cell-inflammatory-genes-in-systemic-lupus/. Accessed .« Back to ACR Convergence 2022
ACR Meeting Abstracts - https://acrabstracts.org/abstract/physical-inactivity-is-associated-with-increased-expression-of-t-cell-inflammatory-genes-in-systemic-lupus/