Session Information
Date: Tuesday, October 28, 2025
Title: (2524–2546) Vasculitis – Non-ANCA-Associated & Related Disorders Poster III
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: The systemic vasculitides are a family of rare diseases defined by immune-mediated inflammation and destruction of vasculature. Causal etiologies of most forms of vasculitis are unknown. Classification schemes to distinguish between forms of vasculitis typically rely on the clinical pattern of disease and do not incorporate biologic data about immune dysregulation. This study aimed to determine the relationship between tissue transcriptomics and clinical diagnoses by profiling skin lesions from patients with different types of vasculitis.
Methods: Patients with skin lesions attributed to one of nine pre-specified vasculitides were recruited into a multi-center prospective observational cohort. Bulk RNA was extracted from 4-6 mm punch biopsies of affected skin using a TRIzol-cholorform method (New England Biolabs Next Ultra II RNA library kit) and sequenced on a Nextseq 2000/NovaseqXPlus (Illumina).
Results: 70 patients underwent punch biopsies of vasculitic skin lesions. The cohort included patients with isolated cutaneous small-vessel vasculitis (CSVV) (n=18), IgA vasculitis (IgAV) (n=15), VEXAS (Vacuoles, E1-enzyme, X-linked, Autoinflammatory, Somatic) syndrome (n=10), granulomatosis with polyangiitis (GPA) (n=7), eosinophilic granulomatosis with polyangiitis (EGPA) (n=6), polyarteritis nodosa (PAN) (n=6), microscopic polyangiitis (MPA) (n=3), urticarial vasculitis (UV) (n=4), or cryoglobulinemic vasculitis (CryoV) (n=1) . The skin lesion morphology included purpuric macules (PM, n = 20), purpuric papules (PP, n=20), urticarial lesions (U, n=10), nodules (N, n=9), plaques (P, n=5), retiform purpura (RP, n=5), and one patient with unknown morphology. Plotting of principal component analysis eigen scores revealed that VEXAS samples clustered independently from the other forms of vasculitis in this cohort (Figure 1). There was no clear clustering based on skin morphology (Figure 2). Differential gene expression analysis revealed a substantial number of significantly differentially expressed genes (DEGs) between VEXAS and the other forms of vasculitis. Gene Ontology enrichment analysis identified a relative upregulation of interferon response genes in VEXAS. The top DEGs that defined this upregulation of the interferon pathway in VEXAS relative to the other forms of vasculitis were XAF1 (log2 fold change = 4.132), ISG15 (log2 fold change = 3.494), and IFIT3 (log2 fold change = 3.453).
Conclusion: Direct profiling of affected tissue across different forms of vasculitis reveals shared and divergent pathophysiology. For example, tissue transcriptomics confirm that VEXAS has a strong interferon signature relative to other types of vasculitis. Clinical assessment of morphology without incorporation of biological data about immune dysregulation misses critical information. Our data supports that future classification schema for systemic vasculitides would benefit from the incorporation of biologic data with clinical features for the characterization of disease. This has the potential to lead to novel and personalized approaches to treatment.
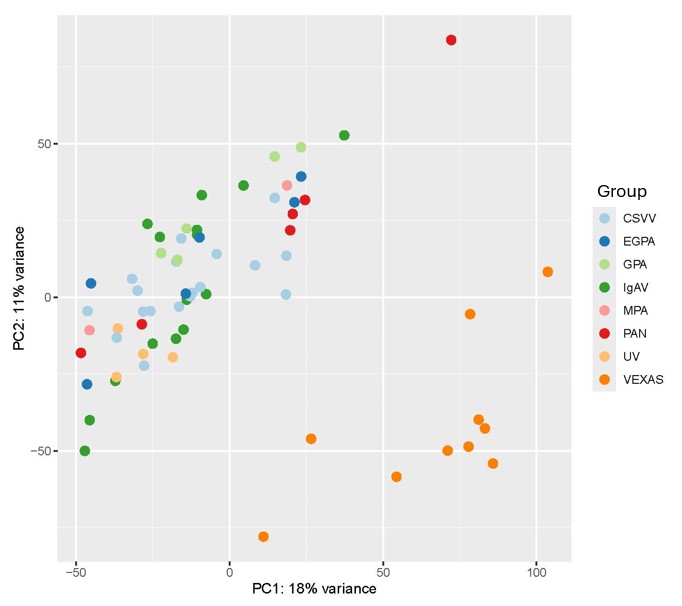 Figure 1: Principal component analysis grouping by type of vasculitis. Patients with cryoglobulinemic vasculitis and outliers were excluded from this analysis.
Figure 1: Principal component analysis grouping by type of vasculitis. Patients with cryoglobulinemic vasculitis and outliers were excluded from this analysis.
PC: Principal component. CSVV: isolated cutaneous small-vessel vasculitis (n=18), EGPA: eosinophilic granulomatosis with polyangiitis (n=6), GPA: granulomatosis with polyangiitis (n=7), IgAV: IgA vasculitis (n=15), MPA: microscopic polyangiitis (n=3), PAN: polyarteritis nodosa (n=6), UV: urticarial vasculitis (n=4), and VEXAS: Vacuoles, E1-enzyme, X-linked, Autoinflammatory, Somatic syndrome (n=10).
.jpg) Figure 2: Principal component analysis grouping by skin lesion morphology among patients with skin manifestations of vasculitis. Patients with cryoglobulinemic vasculitis and outliers were excluded from this analysis.
Figure 2: Principal component analysis grouping by skin lesion morphology among patients with skin manifestations of vasculitis. Patients with cryoglobulinemic vasculitis and outliers were excluded from this analysis.
PC: Principal component. N: nodules (n=9), P: plaques (n=5), PM: purpuric macules (n = 20), PP: purpuric papules (n=20), RP: retiform purpura (n=5), U: urticarial lesions (n=10), and UNK: unknown (n=1).
To cite this abstract in AMA style:
Stonick K, Yamamoto de Almeida L, Jung S, Naz F, Fike A, Quinn K, Banerjee S, Hansen C, Cuthbertson D, Fernandez A, Khalidi N, Kermani T, Langford C, McAlear C, Pagnoux C, Rhee R, Merkel P, Micheletti R, Grayson P. Patterns of skin lesion transcriptomics in different forms of vasculitis [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/patterns-of-skin-lesion-transcriptomics-in-different-forms-of-vasculitis/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/patterns-of-skin-lesion-transcriptomics-in-different-forms-of-vasculitis/
