Session Information
Date: Monday, November 18, 2024
Title: SLE – Treatment Poster III
Session Type: Poster Session C
Session Time: 10:30AM-12:30PM
Background/Purpose: Systemic lupus erythematosus (SLE) is a chronic autoimmune disease with a very heterogeneous clinical presentation and severity. Despite the variety of treatment options available, only about 30% of patients respond to first-line therapy. The integration of systemic lipidomic analyses with other multi-OMIC platforms offers an approach to understand the biological changes that coincide with varying disease activity when using immunomodulatory treatments. Untargeted lipidomic analysis can provide new insights into the characterisation of lipid metabolism, focusing on differences in disease activity status and its changes in response to different immunomodulatory treatments, which have not been previously reported.
Methods: Eleven patients with different activity levels of SLE and induction or continuation of a new DMARD therapy were selected from a prospective observational cohort study. Plasma samples and PBMCs were collected at two visits three months apart. In addition to broad lipidomic profiling, samples were used to characterise systemic immune cell architecture using flow cytometry and targeted proteomics using the OLINK inflammation panel for detailed proteomic analysis. Response to DMARD therapy was assessed based on the difference in the SLEDAI-2K disease activity score between the two visits. A machine learning approach (multiblock sPLS-DA) was used to analyse the variation within the multi-OMIC data in relation to the change in disease activity between the two time points. Correlation structures between cytomic, proteomic and lipidomic analytes were uncovered that were best suited to discriminate the groups.
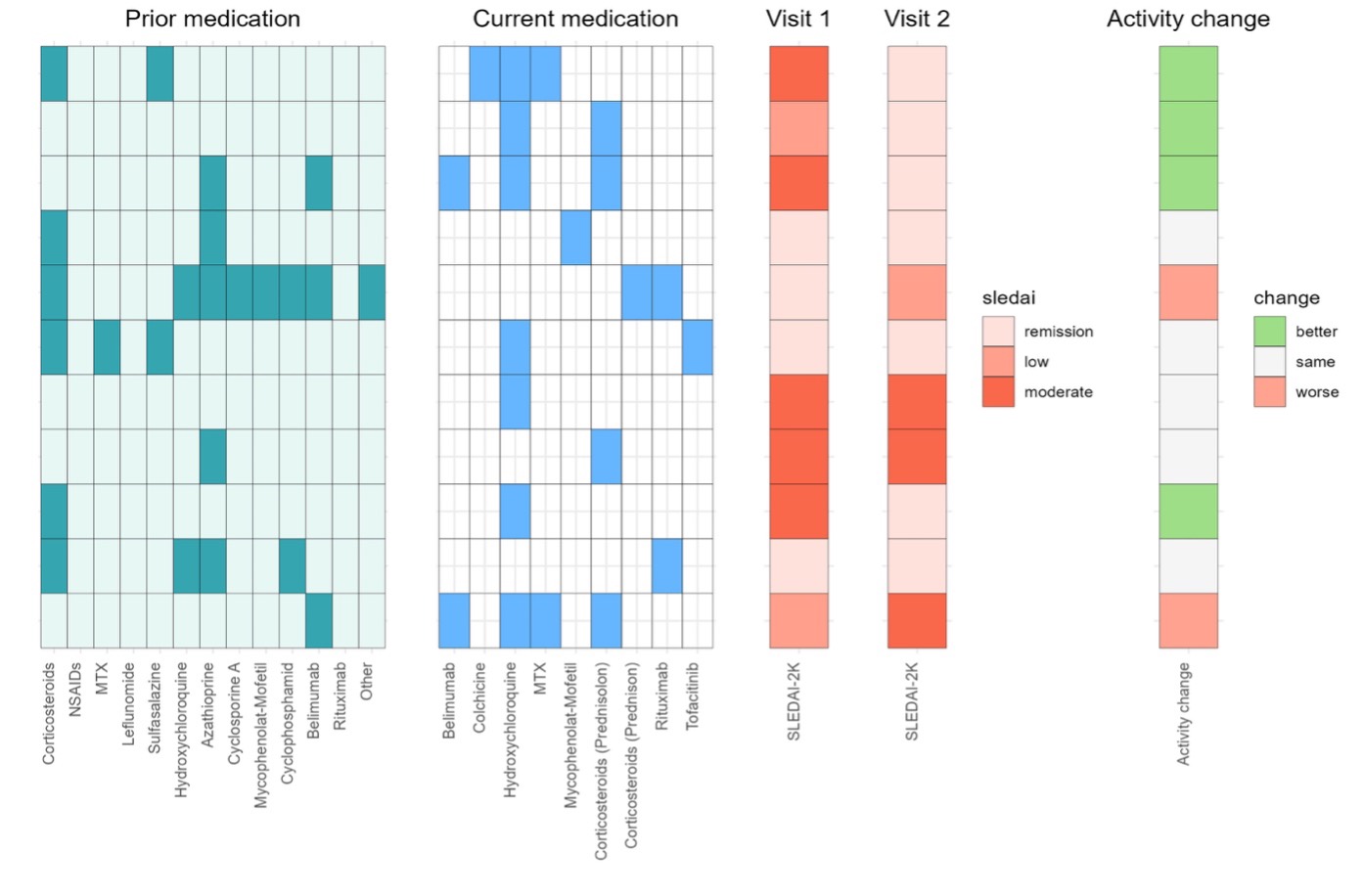
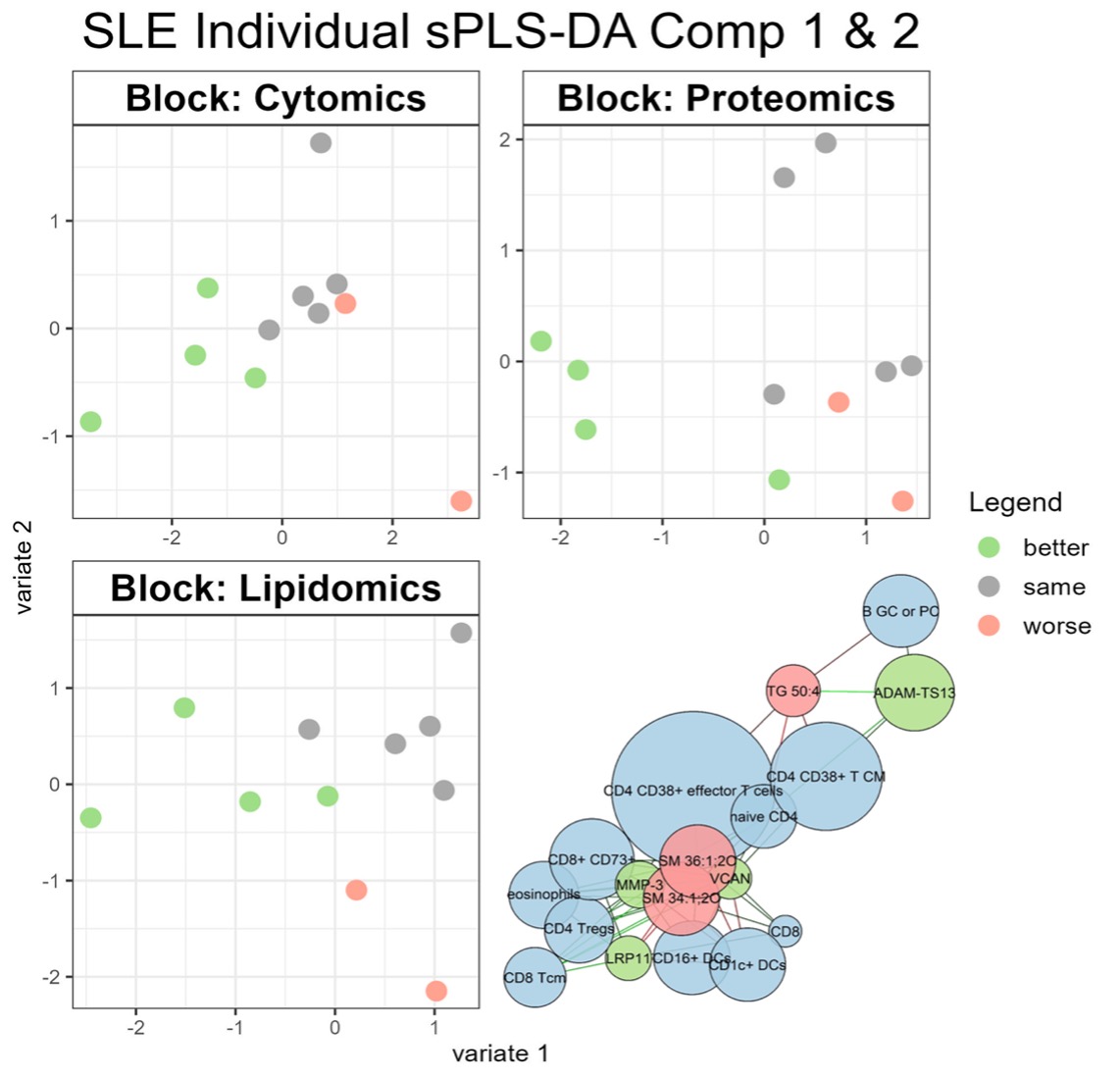
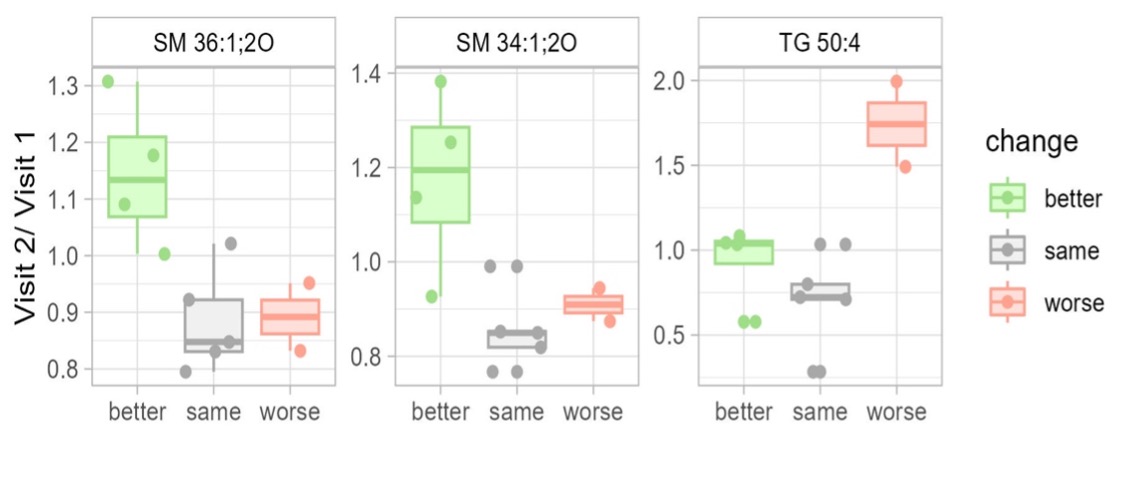
Results: Eleven patients with SLE were selected as an exploratory cohort. According to clinical assessment using the SLEDAI-2K, five patients had moderate disease activity, two had low disease activity and four were in remission at baseline. Pre-treatment and current treatment status with treatment changes are shown in Figure 1A. Despite the small sample size, a supervised machine learning approach (multiblock sPLS-DA) was able to discriminate three disease activity groups based on differences in the systemic cytome, proteome and lipidome between the two visits. Certain correlated features were selected for further investigation (network) (Figure 1B). Two sphingomyelins were selected by the model for optimal discrimination between disease activity related groups based on changes in systemic lipid metabolism (Figure 1C). The two analytes were found to be elevated in patients with improvement in their SLEDAI-2K at the second visit.
Conclusion: This exploratory pilot study was conducted to evaluate the potential of multi-OMICs analysis for predicting disease activity and treatment response in SLE patients. Several studies have suggested that changes in sphingolipid metabolism are associated with changes in SLE disease activity and treatment response, which we can confirm. However, more data are needed to confirm these findings in the future.
To cite this abstract in AMA style:
Koehm M, Rischke S, Gurke R, Geißlinger G, Behrens F. Multi-OMICs Analysis Including Lipidomics to Correlate Baseline OMICs Profiles with Disease Activity and Response to Different Immunomodulatory Treatments in Patients with Systemic Lupus Erythematosus – An Exploratory Pilot Study Using a Multi-OMICs Approach [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/multi-omics-analysis-including-lipidomics-to-correlate-baseline-omics-profiles-with-disease-activity-and-response-to-different-immunomodulatory-treatments-in-patients-with-systemic-lupus-erythematosus/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/multi-omics-analysis-including-lipidomics-to-correlate-baseline-omics-profiles-with-disease-activity-and-response-to-different-immunomodulatory-treatments-in-patients-with-systemic-lupus-erythematosus/