Session Information
Date: Monday, November 11, 2019
Title: 4M095: Spondyloarthritis Including Psoriatic Arthritis – Basic Science (1776–1781)
Session Type: ACR Abstract Session
Session Time: 2:30PM-4:00PM
Background/Purpose: Spondyloarthritis (SpA) results from the interplay between genetic and environmental factors. An emerging factor is the human intestinal microbiota, which multiple studies in children and adults have shown to be abnormal in SpA patients, including enthesitis-related arthritis (ERA) and Ankylosing Spondylitis (AS). Considering the microbiota pathogenic, we may have the opportunity to both identify an at-risk population and consider ways to modify their microbiota. The purpose of this study was to analyze the microbiota of healthy children at risk for SpA, stratified by HLA-B27, and to compare these healthy children to HLA-B27+ ERA patients.
Methods: Children age 5 – 18 years (along with their AS parent) were examined by a rheumatologist for evidence of SpA. Human DNA as well as fecal specimens were collected from both parent and children. Microbiota specimens were subject to amplification of the V4 region of the ribosomal RNA gene, which was sequenced on the Illumina MiSeq device. Host SNP genotyping was performed with the Illumina Global Screening Array, a single nucleotide polymorphism (SNP) microarray with 760,000 markers, from which HLA types were imputed using SNP2HLA. The Quantitative Insight into Microbial Ecology was used for quality control as well as clustering into operational taxonomic units (OTUs) using uclust. The OTU table was imported as a phyloseq object for further processing, visualization, and diversity analysis. DeSeq2 was used for FDR-corrected comparisons of abundances of the major taxa, and the Random Forest algorithm was used to predict the development of arthritis based upon the microbiota contents.
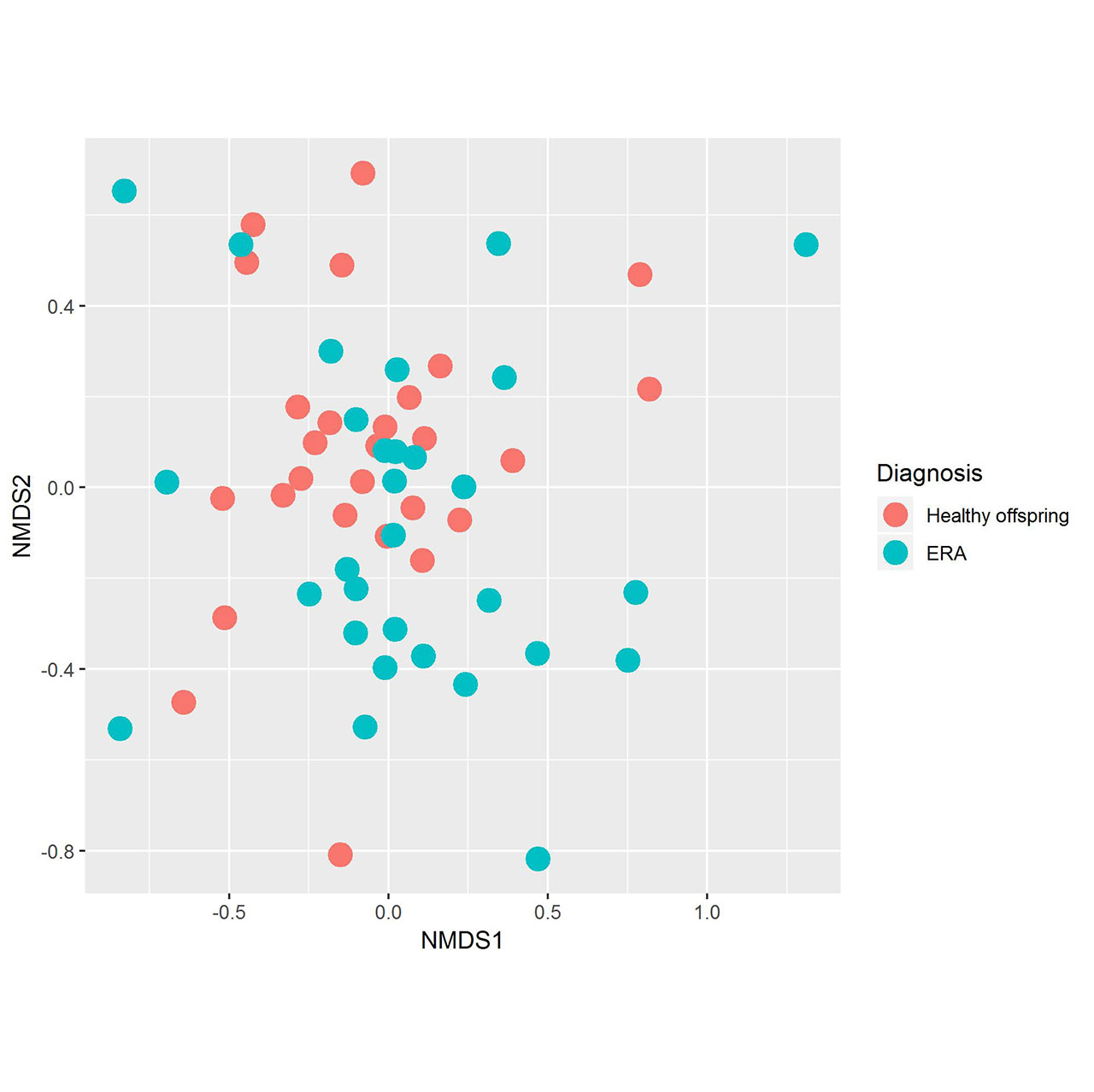
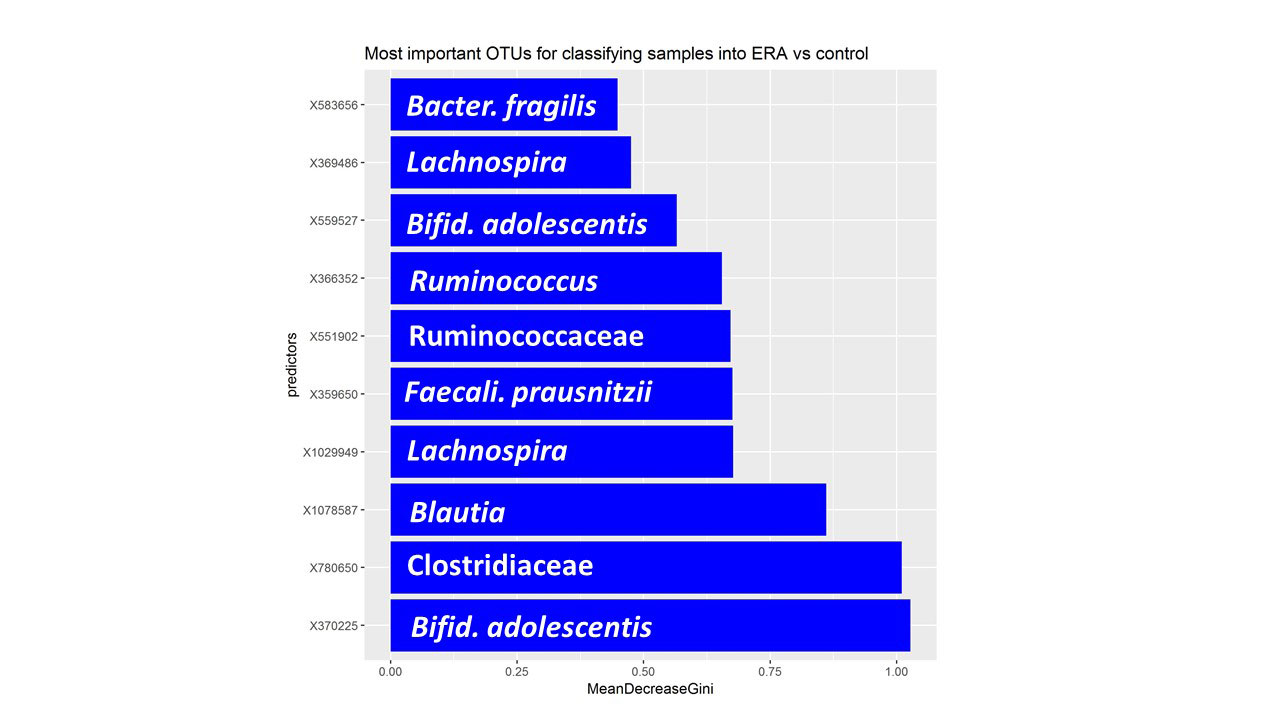
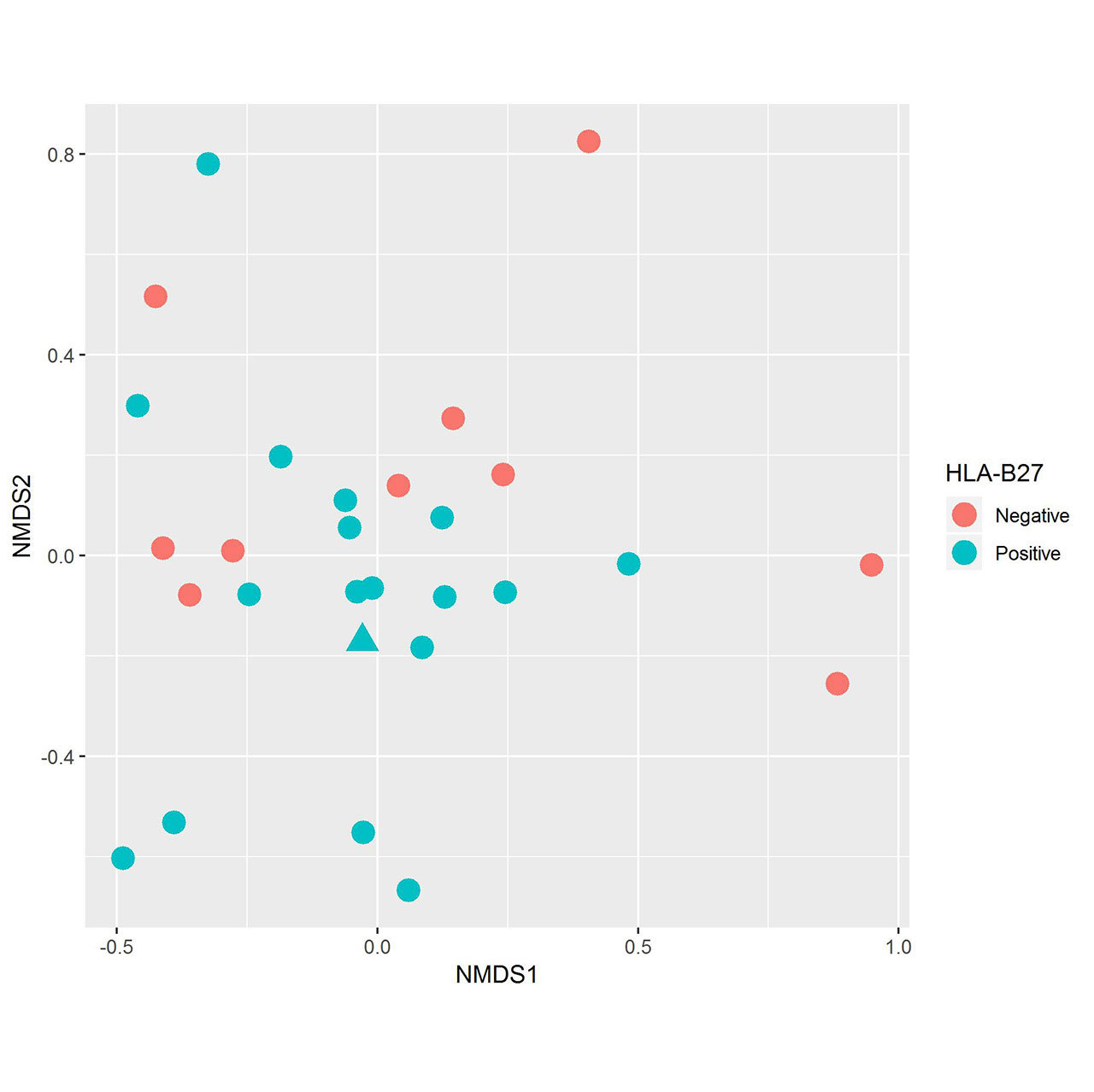
Results: Fecal specimens were obtained on 28 offspring of AS patients, of whom 18 were HLA-B27+; one had a SpA diagnosis at study enrollment. Healthy offspring were compared to 29 additional children with HLA-B27+ ERA (ILAR criteria) recruited from 6 sites. Among ERA patients, clustering by diagnosis was present (Figure 1; p = 0.004, adonis). Following transformations and adjustment for multiple comparisons, 20 OTUs were significantly associated with diagnosis state, of which four (Bifidobacterium adolescentis, Ruminococcaceae, and Clostridiaceae higher in controls; Bacteroides fragilis higher in ERA) were identified among the top 10 most associated with the diagnosis in the Random Forest model (Figure 2). Comparison of the HLA-B27+ to negative offspring revealed clustering by genotype (Figure 3; p = 0.05) and demonstrated that several of the OTUs that distinguished healthy offspring from patients (e.g. Ruminococcus gnavus, higher in ERA vs offspring and HLA-B27+ vs HLA-B27- offspring) were also significantly associated with genotype.
Conclusion: Our data identified several bacteria associated with HLA-B27 in healthy at-risk children and ERA patients, and thus may identify a high-risk population that could be the target of future preventative efforts.
To cite this abstract in AMA style:
Stoll M, DeQuattro K, Li Z, Sawhney H, Castillo M, Weiss P, Nigrovic P, Punaro L, Schikler K, Edelheit B, Reveille J, Brown M, Gensler L. Microbiome in Offspring of Ankylosing Spondylitis Patients [abstract]. Arthritis Rheumatol. 2019; 71 (suppl 10). https://acrabstracts.org/abstract/microbiome-in-offspring-of-ankylosing-spondylitis-patients/. Accessed .« Back to 2019 ACR/ARP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/microbiome-in-offspring-of-ankylosing-spondylitis-patients/