Session Information
Session Type: Poster Session A
Session Time: 10:30AM-12:30PM
Background/Purpose: Sjögren’s disease (SjD) is a systemic autoimmune condition with a complex genetic architecture. To date, 22 genome-wide significant (GWS) SjD risk loci have been identified, while >100 risk loci have been described in other related autoimmune diseases (Khatri et al. 2022). The objective of this study is to perform large-scale genome-wide association study (GWAS) to identify new GWS SjD risk loci (p< 5×10-8; suggestive: p< 5×10-5).
Methods: Institutional IRB/EC approval was obtained. GWAS were performed separately using PLINK2 for individuals of European (EUR) (4,855 cases, 25,408 controls) or East Asian (EAS) (561 cases, 1,760 controls) ancestry classified with SjD. After standard quality control, datasets were phased and imputed using the TOPMed reference panel. Data were meta-analyzed using inverse variance-weighted fixed-effect models in METAL with publicly available biobank GWAS summary statistics for cases [Phecode 709.2 (ICD-10 M35.0, ICD-9 710.2)]: Japanese Biobank (303 EAS cases, 175,599 controls [exclusion criteria Phecodes 690-697.99, 708-709]), UK Biobank (511 EUR cases, 397,694 controls [exclusion criteria Phecodes 690-697.99, 708-709]), Estonian Biobank (464 EUR cases, 204,701 controls [exclusion criteria not-cases]), FinnGen (3,309 EUR cases, 484,260 controls [exclusion criteria ICD-10 M30, M31, M32, M33, M34, M35, M36, and ICD-9 136.1, 446, 447.5, 447.6, 710, 725]).
Results: Thirteen novel GWS risk loci were identified: TNFSF4, IKBKE, IDUA-SLC26A1, IKZF1, NCF1, HIP1, CLDN23-ERI1, ARID5B, PHRF1-IRF7, OVOL1, SH2B3, IRF8, and TMEM86B (Figure 1A and B), with most having been previously implicated in other autoimmune diseases. To investigate the regulatory potential of GWS variants, we analyzed three loci – TNFSF4, IKZF1, and SH2B3 – using FUMA, which integrates chromatin interaction and expression quantitative trait locus (eQTL) data from publicly available immune cell datasets. At the TNFSF4 locus, GWS variants showed both chromatin interactions and eQTLs with RABGAP1L, RC3H1, SERPINC1, ZBTB37, DARS2, CENPL, KLHL20, PRDX6, TNFSF4, TNFSF18, FASLG and SUCO (Figure 1C). At the IKZF1 locus, GWS variants showed both chromatin interactions and eQTLs with FIGNL1, IKZF1 and ZPBP (Figure 1D). At the SH2B3 locus, GWS variants showed both chromatin interactions and an eQTL with PPP1CC (Figure 1E).
Conclusion: By conducting a GWAS using a larger sample size and performing meta-analysis, this study advances the genetic understanding of SjD. Further, SjD risk loci previously established in related autoimmune diseases may be indicative of shared mechanisms of disease. While loci are annotated by the gene closest to the lead variant, this does not necessarily indicate functional relevance. Three loci were evaluated for risk variant impact on chromatin interactions and gene expression changes in immune cells (Figure 1C-D). As both chromatin interactions and eQTLs are often cell-type specific, further analyses across all novel loci and in disease-relevant cell types and tissues are needed. Additional datasets from other SjD clinical partners and biobanks to further increase power are needed to discover novel loci.AcknowledgementsEstonian Biobank Research Team
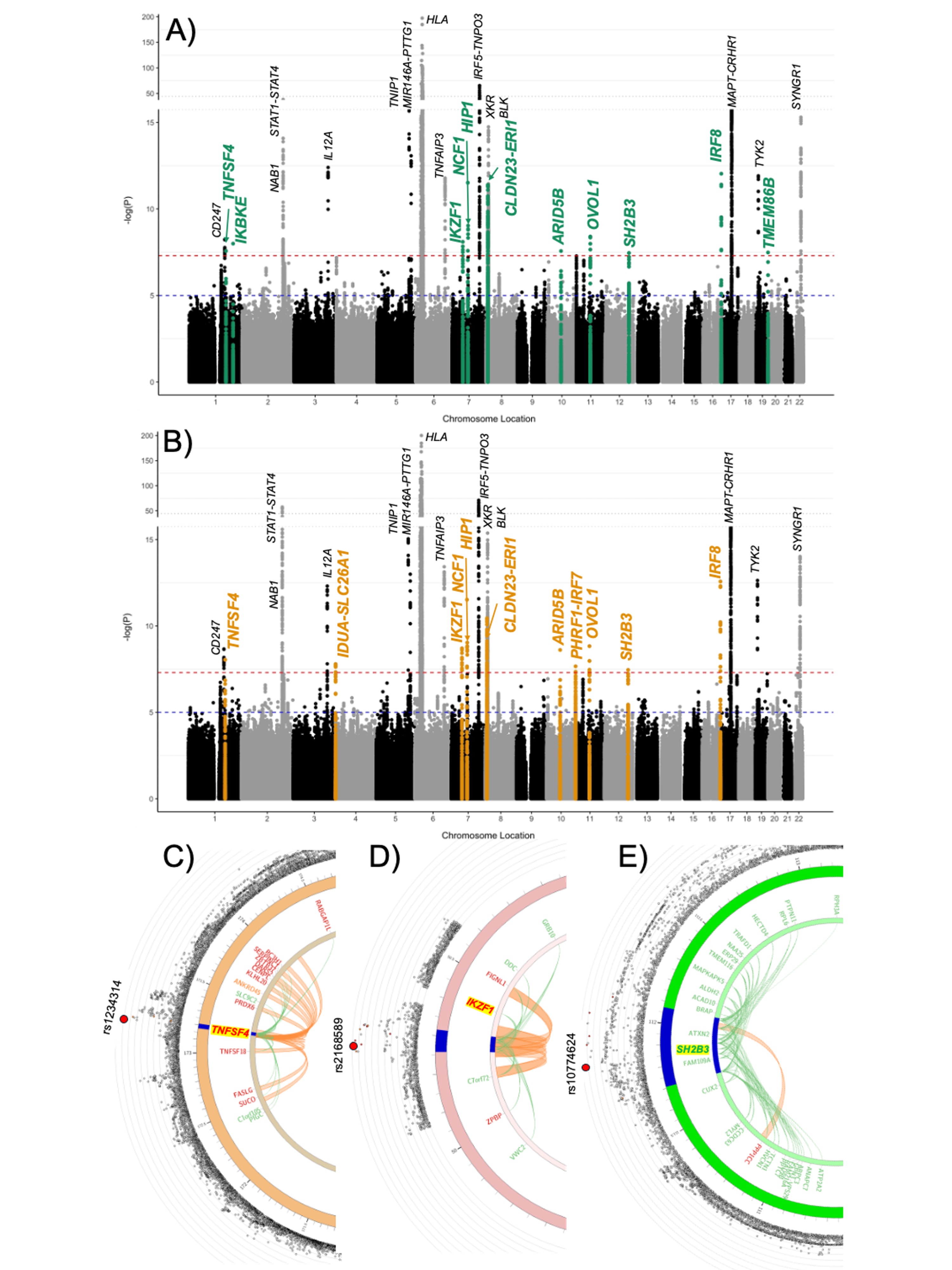 Fig 1. (A-B) The summary statistics of meta-analysis (A) 9,139 EUR cases (green indicates GWS SjD risk loci) or (B) 10,003 EUR+EAS cases (orange indicates GWS SjD risk loci). The -log10(P) for each variant is plotted against genomic locations. Red line indicates GWS (5×10-8) threshold, blue line suggestive threshold (1×10-5). (C-E) Regional maps of the genetic risk locus showing variants associations from the EUR+EAS GWAS shown in B (outer ring; index SNP labeled) and mapped chromatin interactions (orange), eQTLs (green), or both (red) (inner ring) spanning the (C) TNFSF4 (chromosome 1), (D) IKZF1 (chromosome 7), and (E) SH2B3 (chromosome 12) GWS risk loci (indicated as blue in the middle ring).
Fig 1. (A-B) The summary statistics of meta-analysis (A) 9,139 EUR cases (green indicates GWS SjD risk loci) or (B) 10,003 EUR+EAS cases (orange indicates GWS SjD risk loci). The -log10(P) for each variant is plotted against genomic locations. Red line indicates GWS (5×10-8) threshold, blue line suggestive threshold (1×10-5). (C-E) Regional maps of the genetic risk locus showing variants associations from the EUR+EAS GWAS shown in B (outer ring; index SNP labeled) and mapped chromatin interactions (orange), eQTLs (green), or both (red) (inner ring) spanning the (C) TNFSF4 (chromosome 1), (D) IKZF1 (chromosome 7), and (E) SH2B3 (chromosome 12) GWS risk loci (indicated as blue in the middle ring).
M. Radziszewski: None; B. Khatri: None; P. Stuart: None; A. Rasmussen: Clinical Outcomes Solutions, 2, Immunovant Corporation, 2; K. Tessneer: None; C. Pritchett-Frazee: None; M. Pattrick: None; E. Pontarini: None; m. Bombardieri: Amgen, 5, 12, Personal fees, GlaxoSmithKlein(GSK), 5, 12, Personal fees, Janssen, 5, 12, Personal fees, Ono Pharmaceuticals Co. Ltd, 1, 2, UCB, 12, Personal fees; M. Rischmueller: Johnson and Johnson, 12, Servier, Advisory Board, Novartis, 6, 12, Clinical trials; M. Kvarnström: None; T. Witte: None; H. Bootsma: Bristol-Myers Squibb(BMS), 2, 5, Medimmune, 2, Novartis, 2, Roche, 2, 5, Union Chimique Belge, 2; G. Verstappen: argenx, 2; F. Kroese: argenx, 2; A. Vissink: None; S. Pringle: None; A. Tzioufas: None; C. Mavragani: None; A. Baer: Bristol-Myers Squibb(BMS), 2; M. Alarcon-Riquelme: None; J. Martin: None; X. Mariette: Galapagos, 2, GlaxoSmithKlein(GSK), 2, Janssen, 2, Novartis, 2, Ose Pharmaceuticals, 5, Pfizer, 2, UCB, 2; G. Nocturne: None; J. Pers: None; J. GOTTENBERG: AbbVie/Abbott, 6, Bristol-Myers Squibb(BMS), 5, 6, CSL Behring, 6, Eli Lilly, 6, Galapagos, 6, Genzyme, 6, Gilead, 6, Merck/MSD, 6, Novartis, 6, Pfizer, 6, Roche, 6, Sanofi, 6; W. Ng: Abbvie, 2, Argenx, 2, BMS, 2, IQVIA, 2, Janssen, 2, Novartis, 2, Resolve Therapeutics, 2, Sanofi, 2; C. Shiboski: None; K. Taylor: None; L. Criswell: None; B. Warner: Mitobridge, subsidiary of Astellas Bio, 5, Pfizer, Inc., 5; A. Farris: Johnson & Johnson Innovative Medicine, 5; J. James: GlaxoSmithKlein(GSK), 2, Progentec, 5; R. Scofield: IQVIA, 1, Jannsen Pharmaceuticals, 1; J. Guthridge: None; D. Wallace: PPD, 2; S. Venuturupalli: None; M. Brennan: Lipella, 2, MeiraGTx, 2, SUN pharmaceuticals, 2; J. Imgenberg-Kreuz: None; L. Rönnblom: AstraZeneca, 6, Biogen, 2; E. Baecklund: None; M. Eloranta: None; S. Johnsen: None; R. Omdal: None; L. Aqrawi: None; Ø. Palm: None; J. Brun: None; D. Hammenfors: None; M. Jonsson: None; S. Appel: None; S. Bucher: None; H. Forsblad: None; T. Mandl: UCB, 3, P. Eriksson: None; M. Wahren-Herlenius: None; E. Abner: None; T. Esko: None; B. Fisher: Bristol-Myers Squibb (BMS), 2, Galapagos, 5, Janssen, 2, 5, Novartis, 2, Servier, 2, 5; R. Gordon: None; G. Hernandez-Molina: None; A. Lee: Novartis, 6; J. Gudjonsson: None; L. Tsoi: Galderma, 5, Janssen, 5; G. Nordmark: None; C. Lessard: Janssen Global Services, 2, Johnson & Johnson, 2, 5.
To cite this abstract in AMA style:
Radziszewski M, Khatri B, Stuart P, Rasmussen A, Tessneer K, Pritchett-Frazee C, Pattrick M, Pontarini E, Bombardieri m, Rischmueller M, Kvarnström M, Witte T, Bootsma H, Verstappen G, Kroese F, Vissink A, Pringle S, Tzioufas A, Mavragani C, Baer A, Alarcon-Riquelme M, Martin J, Mariette X, Nocturne G, Pers J, GOTTENBERG J, Ng W, Shiboski C, Taylor K, Criswell L, Warner B, Farris A, James J, Scofield R, Guthridge J, Wallace D, Venuturupalli S, Brennan M, Imgenberg-Kreuz J, Rönnblom L, Baecklund E, Eloranta M, Johnsen S, Omdal R, Aqrawi L, Palm Ø, Brun J, Hammenfors D, Jonsson M, Appel S, Bucher S, Forsblad H, Mandl T, Eriksson P, Wahren-Herlenius M, Abner E, Esko T, Fisher B, Gordon R, Hernandez-Molina G, Lee A, Gudjonsson J, Tsoi L, Nordmark G, Lessard, C. Meta-Analysis of GWAS data from 10,003 Sjögren’s Disease Cases Identifies Thirteen Sjögren’s Risk Loci. [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/meta-analysis-of-gwas-data-from-10003-sjogrens-disease-cases-identifies-thirteen-sjogrens-risk-loci/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/meta-analysis-of-gwas-data-from-10003-sjogrens-disease-cases-identifies-thirteen-sjogrens-risk-loci/
