Session Information
Date: Saturday, November 6, 2021
Title: Spondyloarthritis Including PsA – Basic Science Poster (0046–0068)
Session Type: Poster Session A
Session Time: 8:30AM-10:30AM
Background/Purpose: Susceptibility to ankylosing spondylitis (AS) is known to be highly influenced by genetic factors, with heritability assessed in twins of >90%, with >115 genetic variants having been demonstrated to be associated with the disease. Assignment of causality from association studies remains challenging, and for many AS-associated loci the key gene and associated protein involved remain unclear. Using large-scale proteomic datasets and Mendelian randomisation approaches, potential disease-causative associations, where disease-associated genes operate through effects on protein levels to cause the disease concerned, can be investigated. We aimed to identify possible plasma protein level and pathway level changes that may influence the development of AS.
Methods: For the selection of protein exposures, we used an established pipeline (Zheng J et al, 2020). Briefly, this pipeline consisted of five plasma-based pQTL studies of European individuals, in which pQTLs of 1,064 proteins were pooled and used as exposure instruments for MR analysis. We also included a sixth study from a subset of the TwinsUK dataset (n=184, 33 pQTLs) to increase the number of available pQTL exposure instruments in our analysis. To determine AS outcome data for the MR analysis, we used a large-scale GWAS of Caucasian AS cases (n=11,352) vs controls (n=35,446). We performed Mendelian randomisation (MR) using the ‘TwoSampleMR’ package in R v4.0.2 to estimate the putative causal effect of the 1,097 plasma proteins on AS incidence. To ascertain the biological effect of the protein changes, we conducted over-representation analysis (ORA) using the GOBP function of the ‘clusterprofiler’ package in R to determine pathway changes resulting from likely pQTLs.
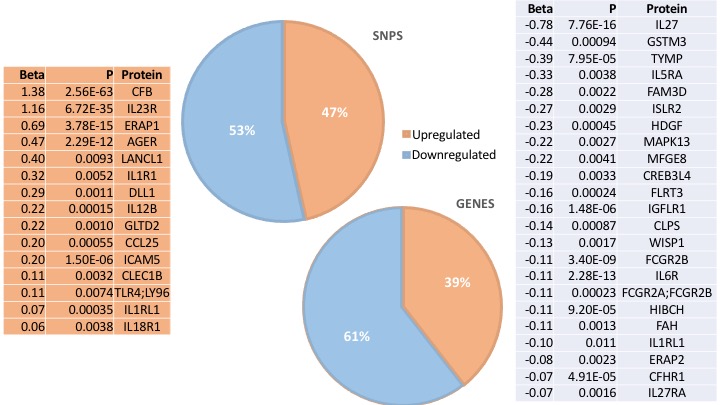
Results: We identified 37 plasma proteins showed putative causal link with AS using MR (FDR< 0.05).
Of these, n=19 (45%) SNPs were likely to increase the effector protein, whereas n=23 (55%) were predicted to have a limiting effect on protein expression. Upregulated proteins included ERAP1 (rs27033, rs17482078, rs469735), IL-23R (rs11581607), IL-12B (rs4921484), and IL-1RL1 (rs10179654). Downregulated proteins included IL-27 (rs181209), GSTM3 (rs115929572), IL-5RA (rs77400868), IL-6R (rs4129267), and ERAP2 (rs17408150). The pQTLs corresponded to immune based gene ontology terms such as ‘GO:0032693 – negative regulation of IL-10 production’, ‘GO:0002697 – regulation of immune effector processes’, and ‘GO:0051707 – response to other organism’.
Conclusion: The results obtained strengthen the hypothesis that immune proteins, particularly cytokines in the IL-23 pathway (including IL-12, IL-23 and IL-27) and their receptor (IL-23R), as well as novel pathways such as the IL-33 pathway, are inherently dysregulated in AS patients. In addition, the ‘response to other organism’ processes may indicate a possible mechanism for gut dysbiosis in AS. These findings point to novel potential therapeutic targets in AS, and support further research targeting proteins upstream of IL-23R for therapy or prevention of AS.
To cite this abstract in AMA style:
Harvey N, Li Z, Garrido-Mesa J, Zheng J, Wordsworth P, Reveille J, Falchi M, Rossi N, Visconti A, Evans D, Brown M. Mendelian Randomisation Analysis of Protein Quantitative Trait Loci with Ankylosing Spondylitis Reveals Causative Involvement of Key Effector Proteins and Pathways [abstract]. Arthritis Rheumatol. 2021; 73 (suppl 9). https://acrabstracts.org/abstract/mendelian-randomisation-analysis-of-protein-quantitative-trait-loci-with-ankylosing-spondylitis-reveals-causative-involvement-of-key-effector-proteins-and-pathways/. Accessed .« Back to ACR Convergence 2021
ACR Meeting Abstracts - https://acrabstracts.org/abstract/mendelian-randomisation-analysis-of-protein-quantitative-trait-loci-with-ankylosing-spondylitis-reveals-causative-involvement-of-key-effector-proteins-and-pathways/