Session Information
Date: Tuesday, November 14, 2023
Title: (1776–1795) Spondyloarthritis Including Psoriatic Arthritis – Basic Science Poster
Session Type: Poster Session C
Session Time: 9:00AM-11:00AM
Background/Purpose: Reactive arthritis (ReA) is the unique infection-triggered spondyloarthritis (SpA). Undifferentiated peripheral SpA (UpSpA) is similar but without previous infection, or psoriasis or inflammatory bowel disease. Around 50-70% of these remit spontaneously while the rest become chronic. There are no known markers of chronicity. We have previously described novel gut-microbiota associated with ReA/UpSpA1. Now, we have attempted to use these microbiota to predict which of these patients would develop chronicity.
Methods: ReA and UpSpA recruited in the previous study1 were followed up at 3 years after the initial recruitment. They were classified as having “Chronic ReA” (still on DMARDs or having symptoms of SpA) or in “Remission” (drug-free and without any symptoms in the last 12 months). A standard pipeline was used to analyze differentially abundant microbiota between Chronic ReA and Remission groups. Further, the groups were randomly divided into training (80% data) and validation (20% data) sets. Clinical features (disease duration, DMARD use, HLA-B27 status) and microbiota of differential abundant families were analyzed by supervised machine learning algorithms (Support Vector Machine: SVM and Linear Regression: LR) to build models that could predict the development of chronicity of ReA.
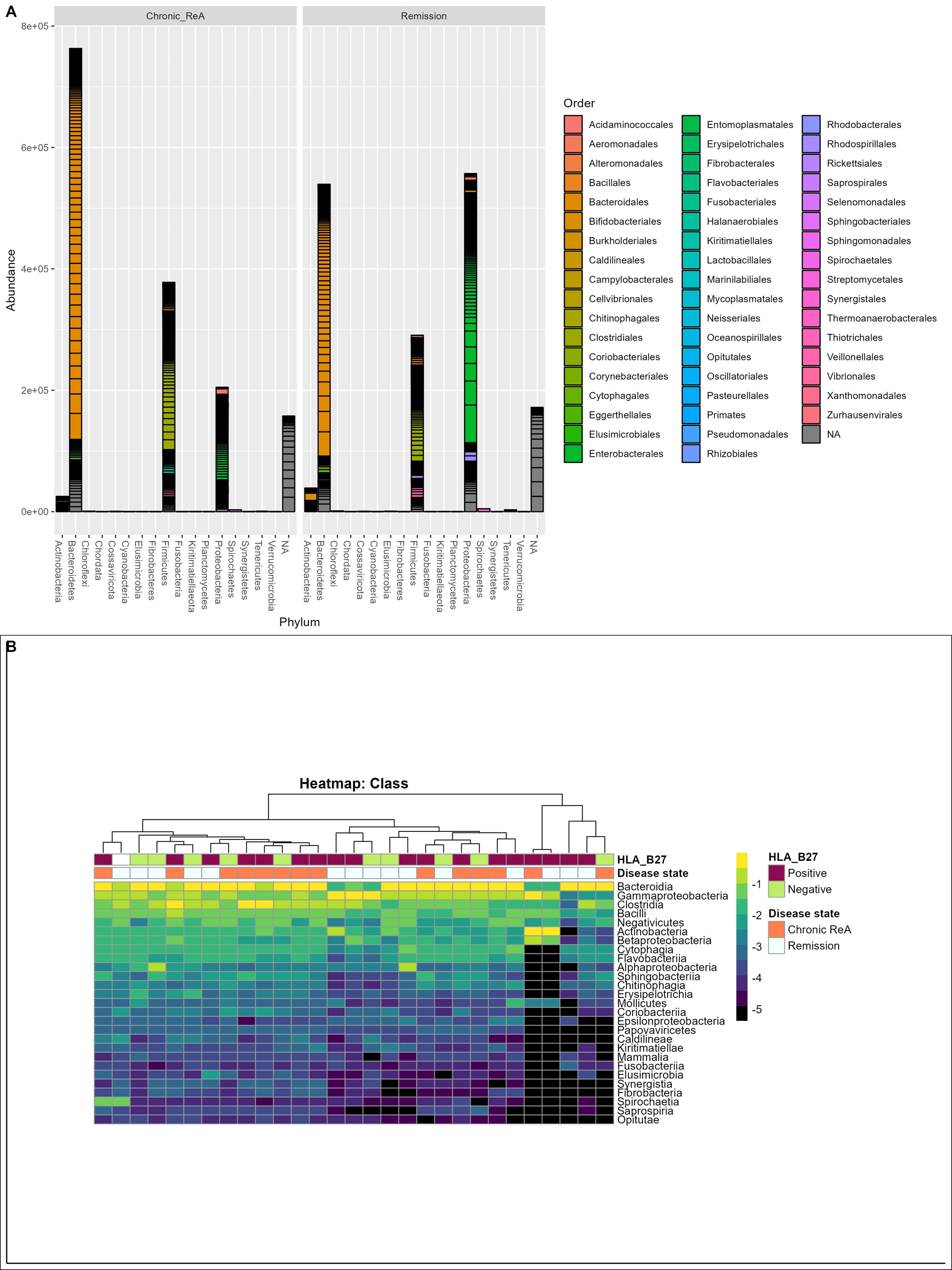
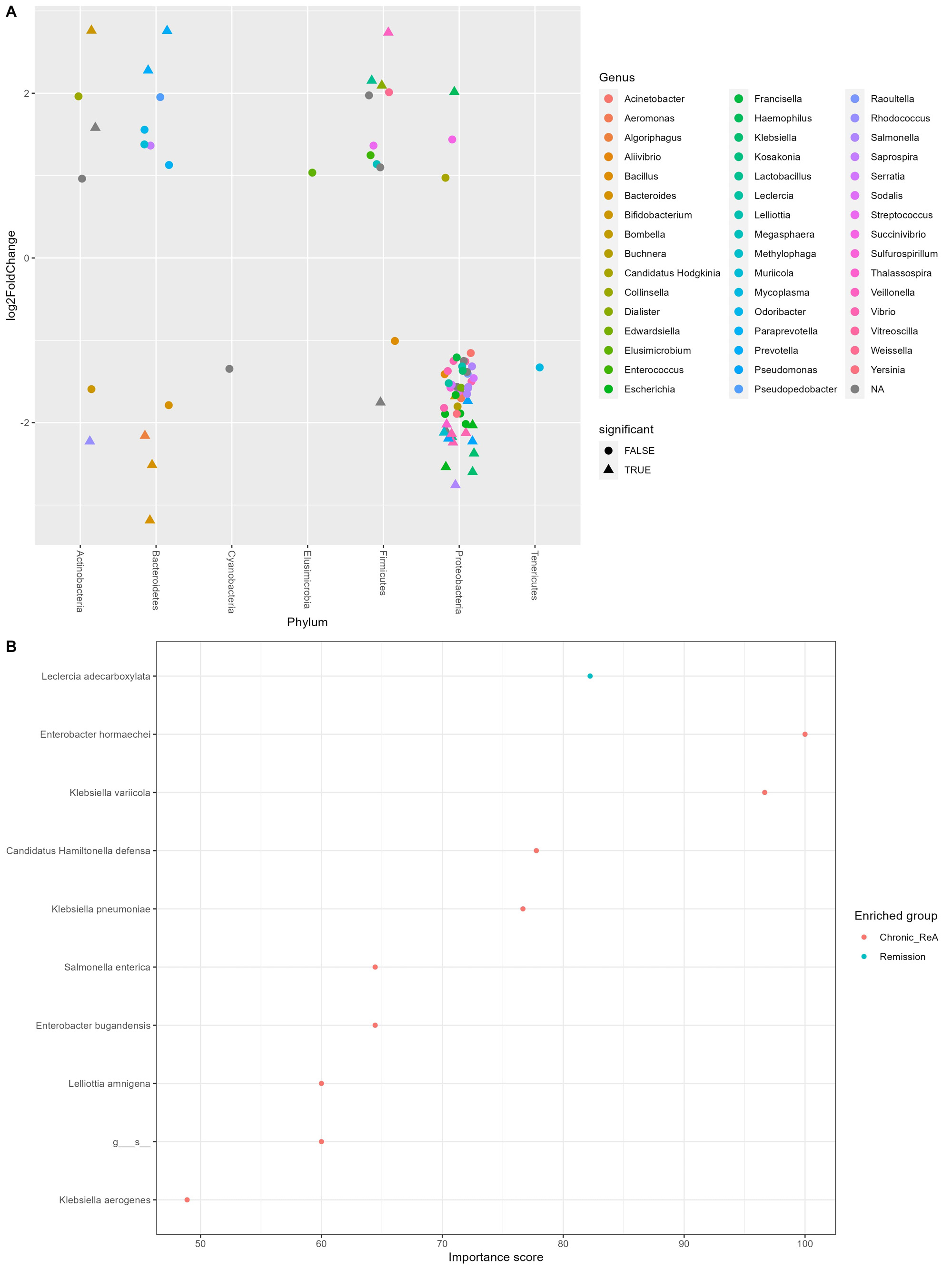
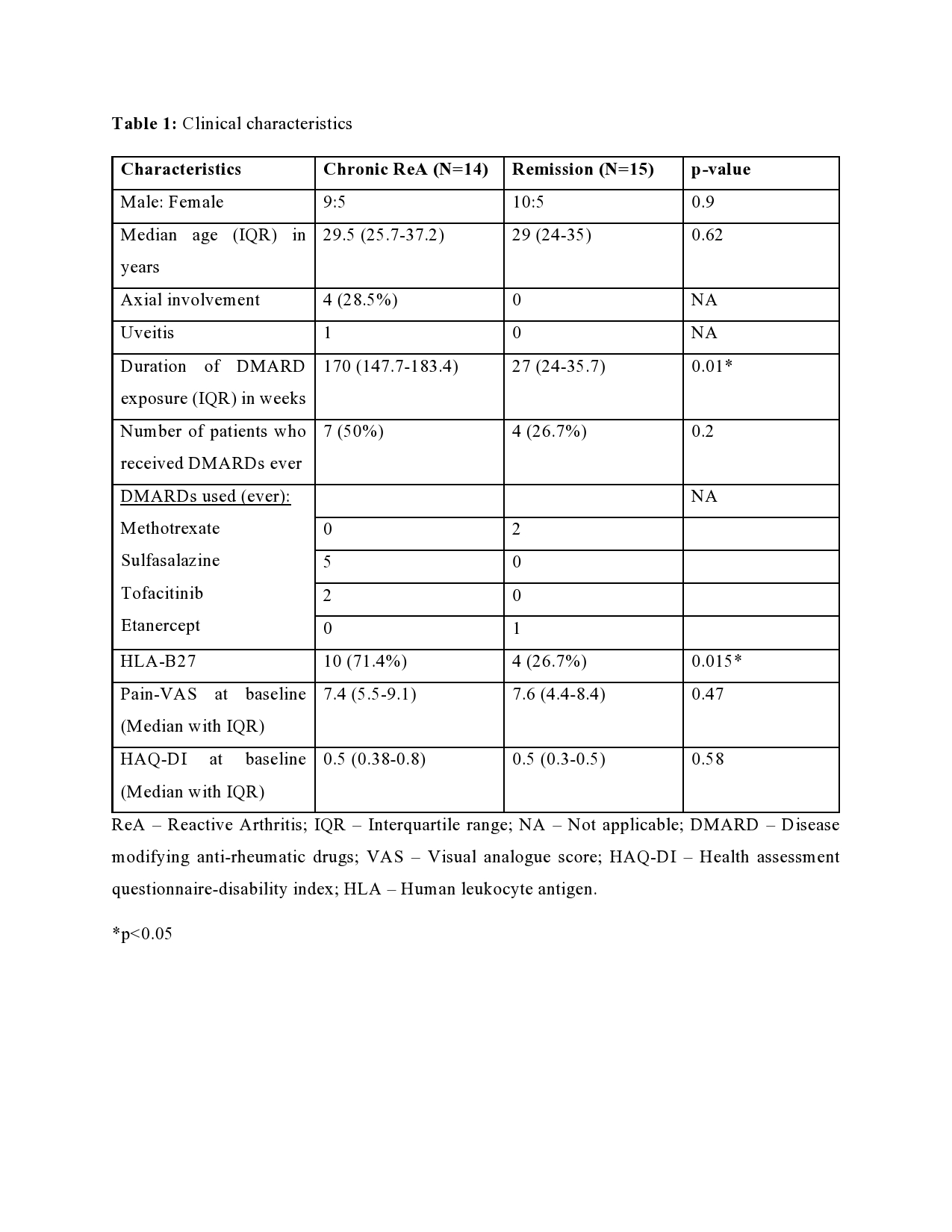
Results: Out of the initial 55 cohort, 29 patients consented for the follow-up. 14 (48.3%) patients had chronic ReA. The clinical features are summarized in Table 1. The baseline microbiome data showed that alpha diversity (Observed, Shannon, Simpson, Fisher, InvSimpson) as well as Beta diversity (Bray-Curtis and Unweighted UniFrac) was similar between the two groups. After applying stringency filters (excluding OTUs that did not appear more than 5 times in least half the samples), the differential abundance is shown at the Phylum (Figure 1A) and Class levels (Figure 1B). Amongst microbiota whose abundance was at least log-2-fold different between Chronic ReA and Remission groups, we could identify 29 OTUs (Figure 2A) that were significantly different (FDR< 0.05). The maximum difference was seen for Bacteroides ovatus (log2fd=3.19), Salmonella sp. HNK130(2.76), Escherichia coli (2.54), Bacteroides fragilis (2.51) that were more abundant in the Chronic ReA group while Veillonella parvula (-2.74), Prevotella dentalis (-2.76), and Bifidobacterium adolescentis (-2.76) were more abundant in the Remission group. In the SVM model, the most accurate model was the one incorporating HLA-B27, the use of methotrexate and of sulfasalazine, and the families Enterobacteriaceae and Bifidobacteriaceae. The validation cohort showed an accuracy of 83% (95% CI: 0.36-0.99). Figure 2B summarizes the species most important for prediction in the LR model.
Conclusion: We have identified microbiota that can predict the chronicity of ReA. However, the accuracy of the prediction can be improved upon with larger cohorts with prospective validation. Reference: < 1. Ahmed S, Mishra R, Mahapatra S, Murmu KC, Padhan P,Prasad P, Punit and Misra R. 16S Metagenomics Reveals Unique Diversity and Novel Gut Microbiota Associated with Reactive Arthritis. Available at: ssrn.com/abstract=4455358.
B – Heatmap visualizing the differential abundance at class level with dendrogram showing the phylogenetic relationship between different samples.
B – Species that are most important for predicting chronicity of Reactive Arthritis as per Linear Regression Machine learning model.
ReA – Reactive Arthritis; IQR – Interquartile range; NA – Not applicable; DMARD – Disease modifying anti-rheumatic drugs; VAS – Visual analogue score; HAQ-DI – Health assessment questionnaire-disability index; HLA – Human leukocyte antigen.
*p<0.05
To cite this abstract in AMA style:
MV P, Mahapatra S, Murmu K, Mishra R, Padhan P, Prasad P, Misra R, Ahmed S. Machine Learning Models Identify Gut Microbiota That Predict Chronicity in Reactive Arthritis [abstract]. Arthritis Rheumatol. 2023; 75 (suppl 9). https://acrabstracts.org/abstract/machine-learning-models-identify-gut-microbiota-that-predict-chronicity-in-reactive-arthritis/. Accessed .« Back to ACR Convergence 2023
ACR Meeting Abstracts - https://acrabstracts.org/abstract/machine-learning-models-identify-gut-microbiota-that-predict-chronicity-in-reactive-arthritis/