Session Information
Session Type: Poster Session B
Session Time: 10:30AM-12:30PM
Background/Purpose: Connective tissue diseases (CTDs) are a family of heterogeneous autoimmune diseases with overlapping clinical features. Not all patients with features suggestive of a mendelian form of lupus or lupus-spectrum diseases, such as childhood-onset disease, are offered genetic testing as part of their rheumatology care. Thus, they do not receive personalised treatment according to their genetic background1. In this multi-centre study, we aimed to investigate if rare pathogenic gene variants in lupus-causing genes are present in an unselected cohort of paediatric and adult patients with lupus and other CTDs.
Methods: The Lupus Extended Autoimmune Phenotype (LEAP) cohort is a UK prospective multicentre study of patients with a diagnosed CTD. The LEAP cohort includes adult and paediatric patients with systemic lupus erythematosus (SLE), primary Sjögren’s syndrome, undifferentiated connective tissue disease (UCTD), systemic sclerosis (SSc), mixed connective tissue disease (MCTD) and idiopathic inflammatory myopathy. For the genetic analysis, we identified patients who are more likely to have a monogenic cause of disease (namely males, those with paediatric onset disease, suggestive clinical features, such as chilblains and cognitive dysfunction, family history, consanguinity or high type I interferon signature). Whole exome sequencing (WES) was performed using the Illumina-Exome 2.0 Plus kit. Variants from 20 age- and ethnicity-matched healthy LEAP controls were filtered out at the start of the analysis. The variants were interpreted according to ACMG guidelines, pathogenicity prediction tools, gnomAD allele frequency and the patient’s phenotype.
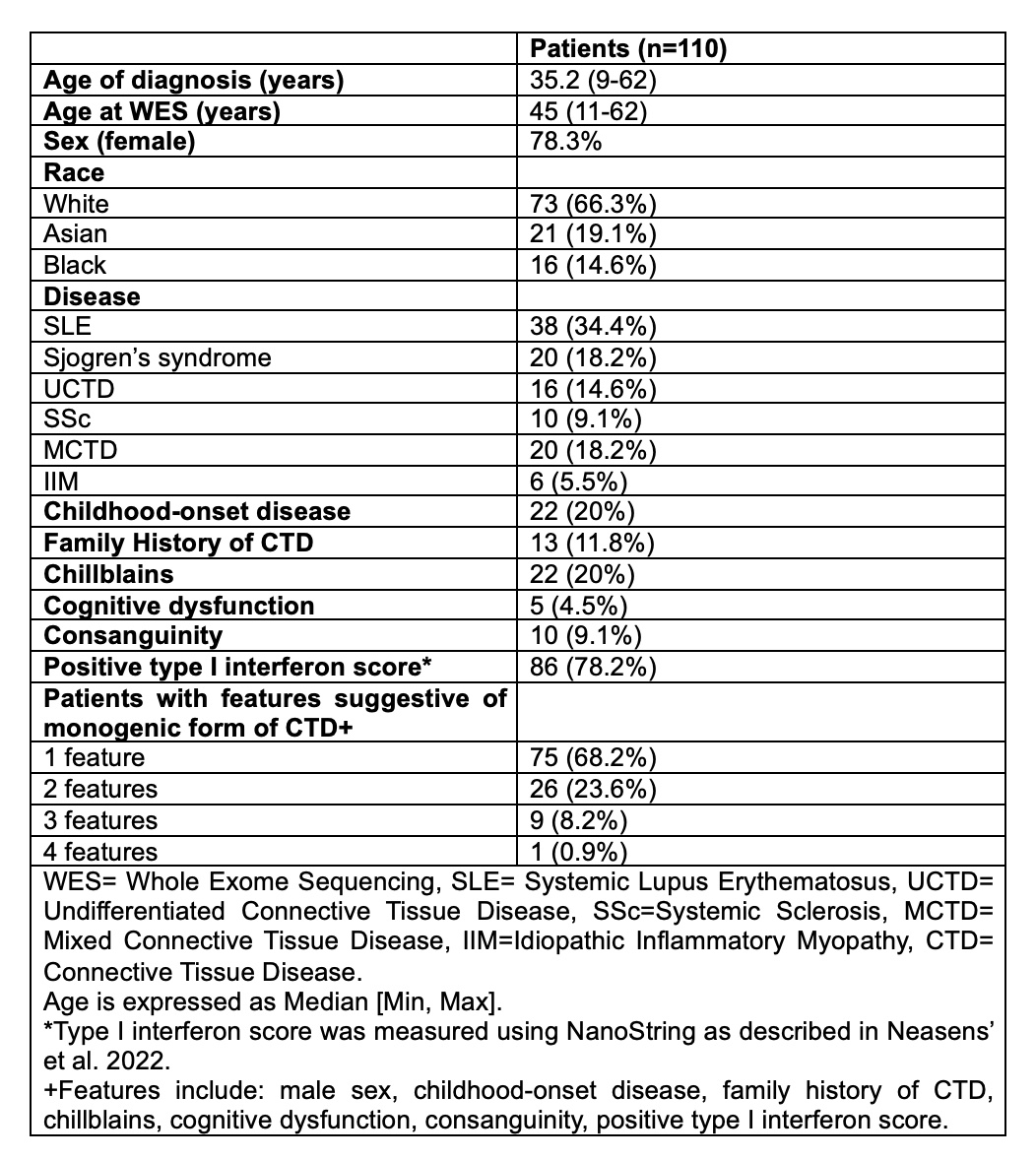
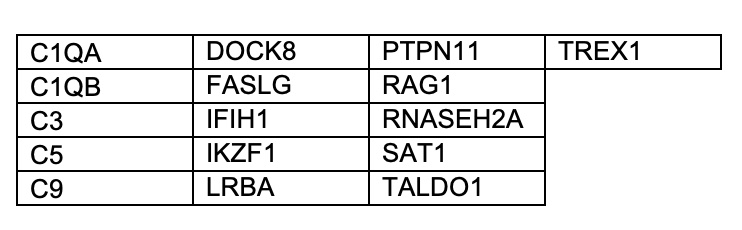
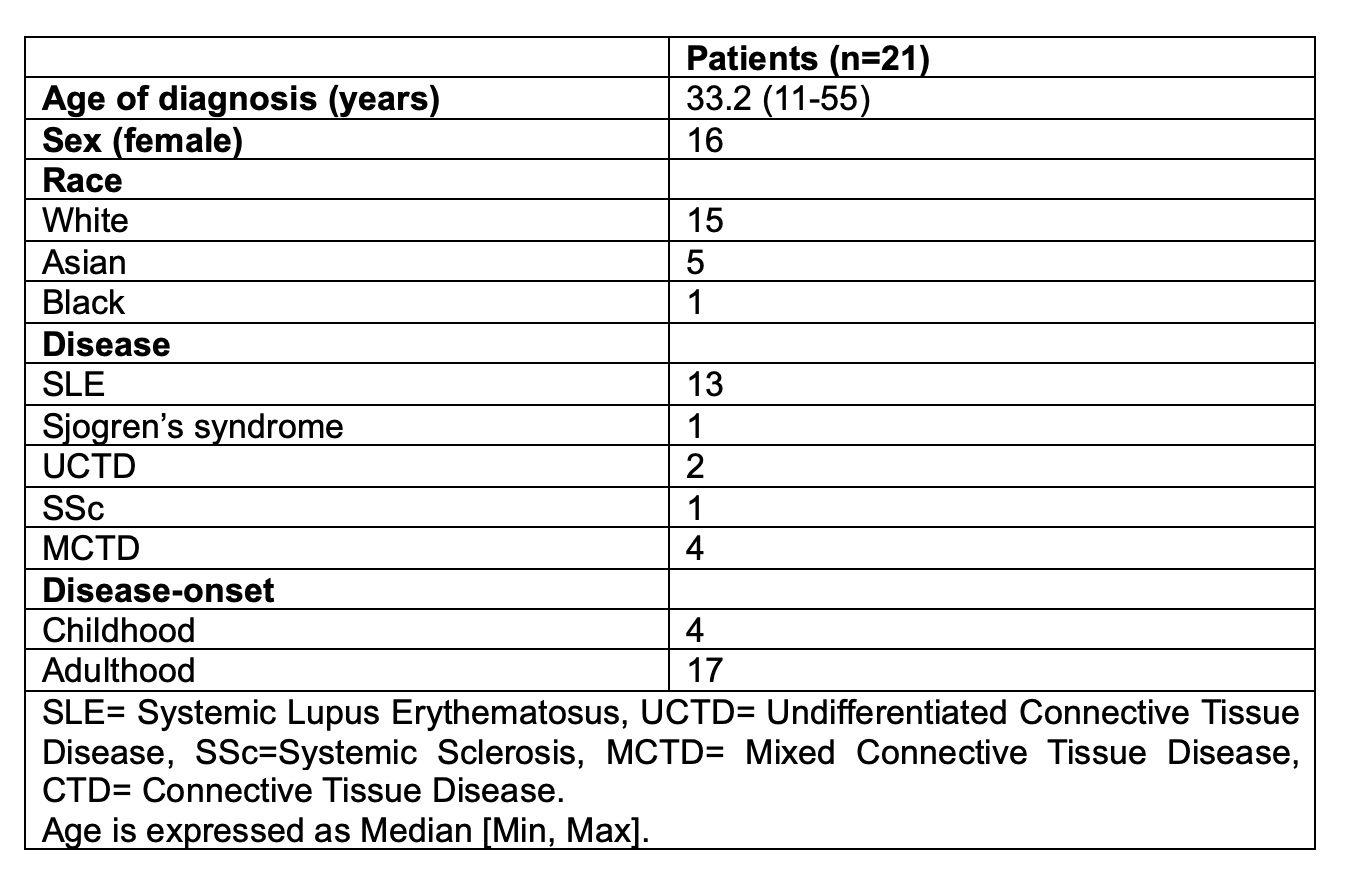
Results: Out of 450 LEAP patients, 110 patients had at least one feature of monogenic disease and underwent WES (Table 1). 21 out of 110 patients (19%) had at least one likely pathogenic missense or splice variant in 16 lupus-causing genes (Table 2). Variants in lupus-causing genes were identified in SLE, UCTD, Sjogren’s and MCTD patients with adult and childhood-onset disease (Table 3). In two more patients, we identified rare missense variants in two genes closely-related to lupus-causing genes waranting further investigation with functional studies.
Conclusion: This study shows that at least 5% (21/450) of patients we see in a CTD clinic may have a monogenic disease, opening up potential for directed therapies, such as off-label use of baricitinib in interferonopathies2. Careful selection of rheumatology patients offered genetic testing can enhance the cost-effectiveness of the test (19% with potentially monogenic disease in our study). To the best of our knowledge, this is the first study indentifying variants in lupus-causing genes in patients with other CTDs suggesting that monogenic forms of disease may exist beyond lupus.
References
1. Briand, C. et al. Efficacy of JAK1/2 inhibition in the treatment of chilblain lupus due to TREX1 deficiency. Ann Rheum Dis 2019.
2. Clinical Commissioning Policy Baricitinib for use in monogenic interferonopathies (adults and children 2 years and over) https://www.england.nhs.uk/wp-content/uploads/2021/07/1930-Baricitinib-policy-Final-1.pdf
To cite this abstract in AMA style:
Madenidou A, Rice G, Garner T, Dyball S, Chieng A, Parker B, Briggs T, Stevens A, Bruce I. Identification of Rare Variants in Lupus-causing Genes in a Mixed Paediatric and Adult Connective Tissue Disease Cohort [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/identification-of-rare-variants-in-lupus-causing-genes-in-a-mixed-paediatric-and-adult-connective-tissue-disease-cohort/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/identification-of-rare-variants-in-lupus-causing-genes-in-a-mixed-paediatric-and-adult-connective-tissue-disease-cohort/