Session Information
Session Type: Abstract Submissions (ACR)
Background/Purpose
Inflammatory bowel diseases (IBD) are associated with changes in microbiota which may be responsible for sustained gut inflammation and/or a consequence of it. Whether variations in microbiota also play a role during the course of inflammatory rheumatic disorders such as SpA or RA, remains to be addressed.
Methods
Targeted metagenomic profiles were established by deeply sequencing the 16S rRNA-encoding bacterial genes amplified by PCR in stools from 97 SpA and 33 RA patients, and 72 healthy controls (including 46 siblings of SpA patients). After pyrosequencing and a denoizing step to remove artefactual reads, bacterial operational taxonomic units (OTUs) were attributed to the remaining sequences (>1.5M reads, 8,000 sequences per sample in average). Supervised analysis (between class co-inertia analysis and L1-regularized logistic machine learning procedure) was carried out to identify factors associated with OTUs variations. Age, gender, disease status and activity (SpA: BASDAI; RA: DAS28) and concomitant treatments (NSAIDs, corticosteroids, DMARDs, biotherapies) were the considered variables.
Results
Neither SpA nor RA status correlated with discrete OTUs variations. Using 600 OTUs represented in at least 1% of the reads mass per sample, disease activity was the main clinical factor explaining variations in OTUs composition, similarly in SpA and RA. Concomitant treatments, and particularly corticosteroids, had also significant impact but of lesser magnitude.
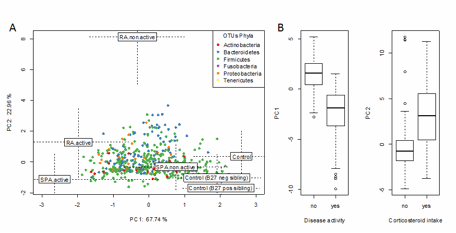
Figure 1: Supervised co-inertia analysis between clinical metadata and microbial OTUs variation as function of health status. A) Health statuses are plotted according the two main components. Monte Carlo test showed that health statuses were significantly linked with clinical metadata and microbiota OTUs variations (colored according their phylum assignation). B) Principal components allowed to significantly separate patients according to disease activity and corticosteroid intake.
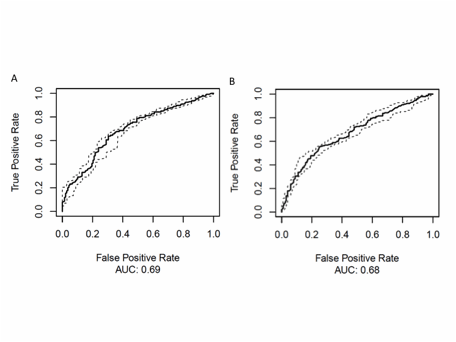
Figure 2: A. Complementary to this signal, SpA and RA could be distinguished from controls by combining 16 OTUs, yielding a statistically significant ROC curve (AUC = 0.69). B. The best model to predict disease activity included 110 OTUs and yielded a statistically significant ROC curve (AUC = 0.68).
Conclusion
Variations in OTUs were found both in SpA and RA, as compared to healthy controls that primarily correlated with disease activity. Whether such variations are cause or consequence of chronic joint inflammation remains to be determined.
Disclosure:
J. Tap,
None;
J. Abou-Ghantous,
None;
A. Leboime,
None;
R. Said Nahal,
None;
P. Langella,
None;
H. J. Garchon,
None;
G. Chiocchia,
None;
J. P. Furet,
None;
M. Breban,
None.
« Back to 2014 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/gut-microbiota-variations-correlate-with-disease-activity-in-spondyloarthritis-spa-and-rheumatoid-arthritis-ra/