Session Information
The 2020 Pediatric Rheumatology Symposium, originally scheduled for April 29 – May 2, was postponed due to COVID-19; therefore, abstracts were not presented as scheduled.
Date: Friday, May 1, 2020
Title: Poster Session 2
Session Type: ACR Abstract Session
Session Time: 5:00PM-6:00PM
Background/Purpose: The genetic contribution to the development of systemic lupus erythematosus (SLE) is estimated to be 66% in twin studies. Genome wide association studies (GWAS) have identified more than 90 SNPs associated with SLE risk. However, there may be additional loci independently impacting the age of diagnosis. The purpose of this study is to identify the genetic variants associated with the age of childhood-onset SLE (cSLE) diagnosis.
Methods: Our cohort included patients with cSLE (diagnosed < 18 years) who met the ACR and/or SLICC classification criteria for SLE, and were followed in tertiary care centers. Patients were genotyped in the Illumina Multiethnic Array (MEGA), and ungenotyped SNPs were imputed using the 1000 genomes project (1KGP) as a reference. We restricted to SNPs with a Minor Allele Frequency (MAF) ≥ 0.01 and imputation quality ≥ 0.8. Principal components (PCs) were calculated, and ancestry was genetically inferred in reference to 1KG. Non-HLA, additive SLE weighted genetic risk scores (GRSs) were computed using published SLE GWAS log-odds ratio weights. Single-variant association tests were performed with GENESIS, a package for accurate ancestry inference that allows us to include patients with admixed ancestry to the analysis. Multivariate linear regression, additive genetic models of age at diagnosis (years) were run genome-wide, adjusting for sex, 5 PCs and SLE non-HLA GRS.
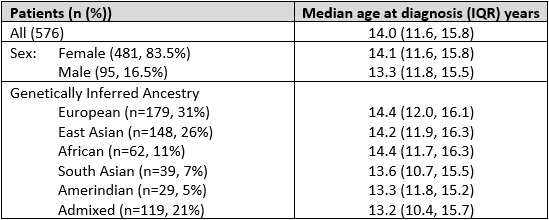
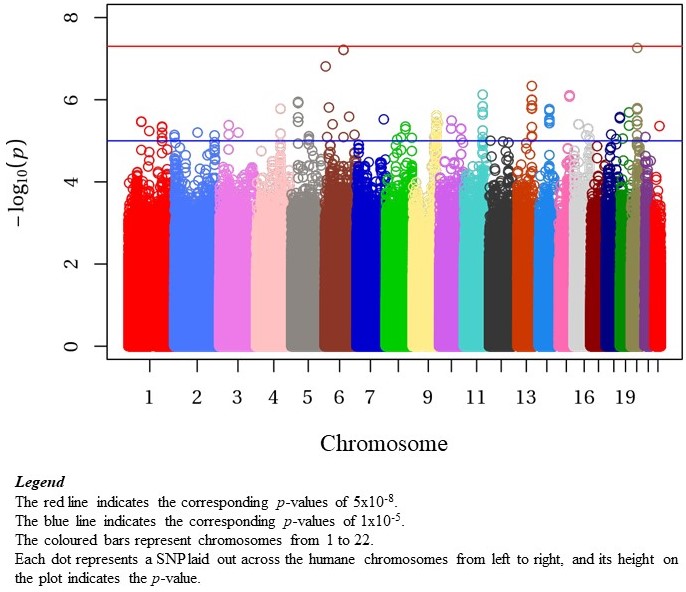
Results: Our cohort included 576 patients, 415 (72%) from The Hospital for Sick Children in Toronto, 115 (20%) from Toronto Western Hospital, and 46 (8%) from Lurie Children’s Hospital of Chicago. The median age at diagnosis was 14.0y (IQR 11.6, 15.8; Range 2.1-17.9y), 83.5% were female, with 31% European and 26% East Asian (Table 1). Genotyping and imputation resulted in 8.7M SNPs included in GWAS. The most significant SNPs associated with age of cSLE diagnosis were on chr6, rs77661956 (Beta=4.43, SE=0.82, p=6.13×10-8, MAF=0.01), and on chr20, rs2425070 (Beta=3.77, SE=0.69, p=5.48×10-8, MAF=0.02) (Figure 1). The chr6 SNP rs77661956 is intronic to ASCC3, a gene encoding helicases implicated in DNA repair. Chr20 SNP rs2425070 is intronic to the NFS1 gene that encodes an aminotransferase required in cellular enzymatic functions.
Conclusion: In our multiethnic cSLE cohort, GWAS did not identify a genome-wide significant SNP association with the age of diagnosis. This could be a result of the relatively modest sample size. There were 2 loci on Chr6 and Chr20, near genome-wide significance. Neither of these loci have been identified in prior GWAS of SLE. Future studies will aim to replicate these signals, as well as explore non-linear associations with age of cSLE diagnosis.
To cite this abstract in AMA style:
Carlomagno R, Liao F, Cao J, Gladman D, Klein-Gitelman M, Knight A, Levy D, Paterson A, Touma Z, Urowitz M, Webber D, Wither J, Silverman E, Hiraki L. Genetics of Age at Diagnosis in Childhood-Onset Systemic Lupus Erythematosus [abstract]. Arthritis Rheumatol. 2020; 72 (suppl 4). https://acrabstracts.org/abstract/genetics-of-age-at-diagnosis-in-childhood-onset-systemic-lupus-erythematosus/. Accessed .« Back to 2020 Pediatric Rheumatology Symposium
ACR Meeting Abstracts - https://acrabstracts.org/abstract/genetics-of-age-at-diagnosis-in-childhood-onset-systemic-lupus-erythematosus/