Session Information
Session Type: ACR Poster Session A
Session Time: 9:00AM-11:00AM
Background/Purpose: Lupus nephritis (LN) is a severe consequence of systemic lupus erythematosus (SLE) that affects minority populations in higher proportions. The causes behind these ethnic differences are not fully elucidated. We aimed to identify genetic variants that contribute differentially to the risk of LN across different ancestral groups.
Methods: 1244 SLE patients from five different racial/ethnic groups were studied. Genome-wide single nucleotide polymorphism (SNP) genotyping was performed with the Axiom LAT1 array (World Array 4, Affymetrix), which is composed of 817,810 SNP markers across the genome and was specifically designed to provide maximal coverage for diverse ethnic populations, including West Africans, Europeans and Native Americans. Genetic ancestry was determined by principal component analysis (PCA). The first 3 PCs (pc1, pc2, pc3) separated Europeans (n=535), Hispanics (n=252), African Americans (n=232) and Asians (n=223). A further PCA was performed to separate northern (n=295) vs southern Europeans (n=242). We utilized a candidate gene approach for genetic variants that have been identified in previous genetic studies of SLE risk, chronic kidney disease and/or LN. Allele frequencies for risk alleles were calculated for each ancestry group. Case-control associations between candidate SNPs and LN were performed within each ancestry group, adjusting for disease duration and principal components. We also utilized meta-analysis to test for heterogeneity and examine associations across the 5 ethnic groups.
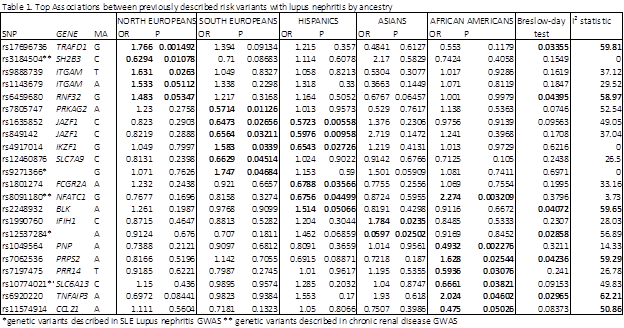
Results: Overall, 606 participants (48.7%) had LN as defined by the ACR renal criterion. The prevalence of LN varied across the ethnic groups, ranging from 38% to 60.7%. Sixty out of 67 candidate gene SNPs varied significantly in frequency across the 5 ancestry groups (p < 7 E-4) in patients without LN. Association tests for risk of LN within each ancestry group revealed differences in the magnitude and significance (Table 1). Moreover, most SNPs were associated with LN for only 1 or 2 ancestry groups. The most significantly associated SNPs included recently identified risk variants for chronic kidney disease GWAS such as NFATC1, suggesting pleiotropy/poligenicity. Tests of heterogeneity (Breslow-day test, I2 square statistic) revealed significant heterogeneity according to ancestry for 6 candidate SNPs, indicating differential contribution of genetic risk variants to risk of LN according to ancestry.
Conclusion: These results demonstrate significant variation in the frequency of risk alleles as well as differential association with the risk of LN according to ancestry. These findings further support the hypothesis that genetic factors contribute importantly to observed ancestry differences in severe outcomes of SLE.
To cite this abstract in AMA style:
Lanata C, Taylor K, Nitiham J, Torgerson D, Tsao BP, Morand EF, Alarcon-Riquelme M, Criswell LA. Genetic Heterogeneity in the Risk of Lupus Nephritis According to Ancestry [abstract]. Arthritis Rheumatol. 2016; 68 (suppl 10). https://acrabstracts.org/abstract/genetic-heterogeneity-in-the-risk-of-lupus-nephritis-according-to-ancestry/. Accessed .« Back to 2016 ACR/ARHP Annual Meeting
ACR Meeting Abstracts - https://acrabstracts.org/abstract/genetic-heterogeneity-in-the-risk-of-lupus-nephritis-according-to-ancestry/