Session Information
Date: Monday, October 27, 2025
Title: (1517–1552) Systemic Lupus Erythematosus – Treatment Poster II
Session Type: Poster Session B
Session Time: 10:30AM-12:30PM
Background/Purpose: Systemic lupus erythematosus (SLE), have a preclinical phase where individuals display a subset of symptoms, but do not meet the full diagnostic criteria. Identifying genetic variants influencing the progression from preclinical stage is crucial, since autoimmunity can quickly cause irreversible organ damage. Biobanks with electronic health records are valuable for studying disease progression but can be underpowered due to limited number of diseases cases. We previously developed genetic prediction scores (GPS, Wang, Markus et al, Nature Communications, 2025) that can predict the progression of SLE from preclinical stages. Compared to conventional genetic risk scores calculated from SLE case-control studies, the GPS method increases the accuracy of risk prediction by ~20%-1000%. Despite the progress, individual variants associated with SLE progressions remain elusive. More powerful methods are needed to understand the etiology of preclinical SLE.
Methods: Almost SLE cases will undergo a preclinical stage with positive antinuclear antibody. Leveraging this insight, we can break the genetic effects influencing healthy → SLE transition into the effects influencing healthy → preclinical and that influencing preclinical → SLE transition, leading a novel Bayesian model to detect genetic association with progression (GAP). GAP borrows strength from the large sample sizes of SLE case-control studies and improves power of GWAS for progression phenotypes (Fig 1). We also developed a novel drug repurposing pipeline that leverages GAP results to identify compounds capable of slowing down the preclinical → SLE progression by reversing the expression levels of causal genes in disease-relevant cell types including B cells.
Results: We apply GAP to identify genetic associations with preclinical → SLE progressions, integrating BioVU biobank (749 SLE cases, 2,784 preclinical individuals, 46,174 controls) and case-control GWAS summary statistics from six different studies (14,355 cases and 505,956 controls). We identified 4 and 17 novel loci for control → preclinical and preclinical → SLE progressions (Fig 2). Intriguingly, two causal variants in IRF5 are colocalized with eQTLs from different genes, emphasizing the importance of resolving phenotype heterogeneities due to different disease stages. Leveraging our drug-repurposing pipeline, we identify several novel drugs, leukotriene receptor antagonist (montelukast) and TNF-α inhibitors (pentoxifylline and lenalidomide). We also confirmed the effectiveness of hydroxychloroquine for slowing down SLE progression, a drug that is frequently prescribed but lacking solid scientific support.
Conclusion: Our results reveal distinct genetic architecture of healthy → preclinical and preclinical → SLE progressions. The novel genes and putative drugs we identify will inform clinical studies and accelerate the therapeutic development for slowing the progression of SLE from preclinical stages.
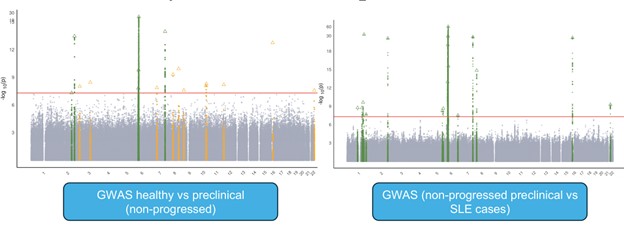 Fig 1. Study design and the key model for the method to detect genetic associations with progression phenotype. Panel A: each SLE patients will undergo a preclinical stage. Panel B. We will decompose the genetic effects into effects influencing healthy to preclinical progressions and the effects influencing preclinical to SLE progressions.
Fig 1. Study design and the key model for the method to detect genetic associations with progression phenotype. Panel A: each SLE patients will undergo a preclinical stage. Panel B. We will decompose the genetic effects into effects influencing healthy to preclinical progressions and the effects influencing preclinical to SLE progressions.
.jpg) Fig 2. GWAS identifies distinct loci influencing different stages of SLE progression. Left panel: GWAS hits influencing healthy to preclinical stage progressions. Right panel: GWAS hits influencing preclinical to SLE progressions. Green color shows loci identified previously from case-control studies and yellow color shows loci unique to SLE progression traits.
Fig 2. GWAS identifies distinct loci influencing different stages of SLE progression. Left panel: GWAS hits influencing healthy to preclinical stage progressions. Right panel: GWAS hits influencing preclinical to SLE progressions. Green color shows loci identified previously from case-control studies and yellow color shows loci unique to SLE progression traits.
To cite this abstract in AMA style:
Wang L, Markus h, Carrel L, Olsen N, Foulke G, Liu D. Genetic architecture and translational insights for SLE progression from preclinical stages [abstract]. Arthritis Rheumatol. 2025; 77 (suppl 9). https://acrabstracts.org/abstract/genetic-architecture-and-translational-insights-for-sle-progression-from-preclinical-stages/. Accessed .« Back to ACR Convergence 2025
ACR Meeting Abstracts - https://acrabstracts.org/abstract/genetic-architecture-and-translational-insights-for-sle-progression-from-preclinical-stages/
