Session Information
Session Type: Poster Session A
Session Time: 10:30AM-12:30PM
Background/Purpose: Electronic health record (EHR) data provide a widely available, inexpensive, and information-rich tool that is underutilized in the research of rare diseases like antiphospholipid syndrome (APS). However, due to the relative complexity and time intensity of classifying APS, as well as likely inaccuracies in EHR coding, it can be challenging to identify high-fidelity cohorts of APS patients that can serve as a starting point for retrospective and prospective clinical research. We present the first computable phenotype for identifying APS patients from EHR data and hope to use this as a platform for identifying collaborators interested in expanding this work to multiple sites.
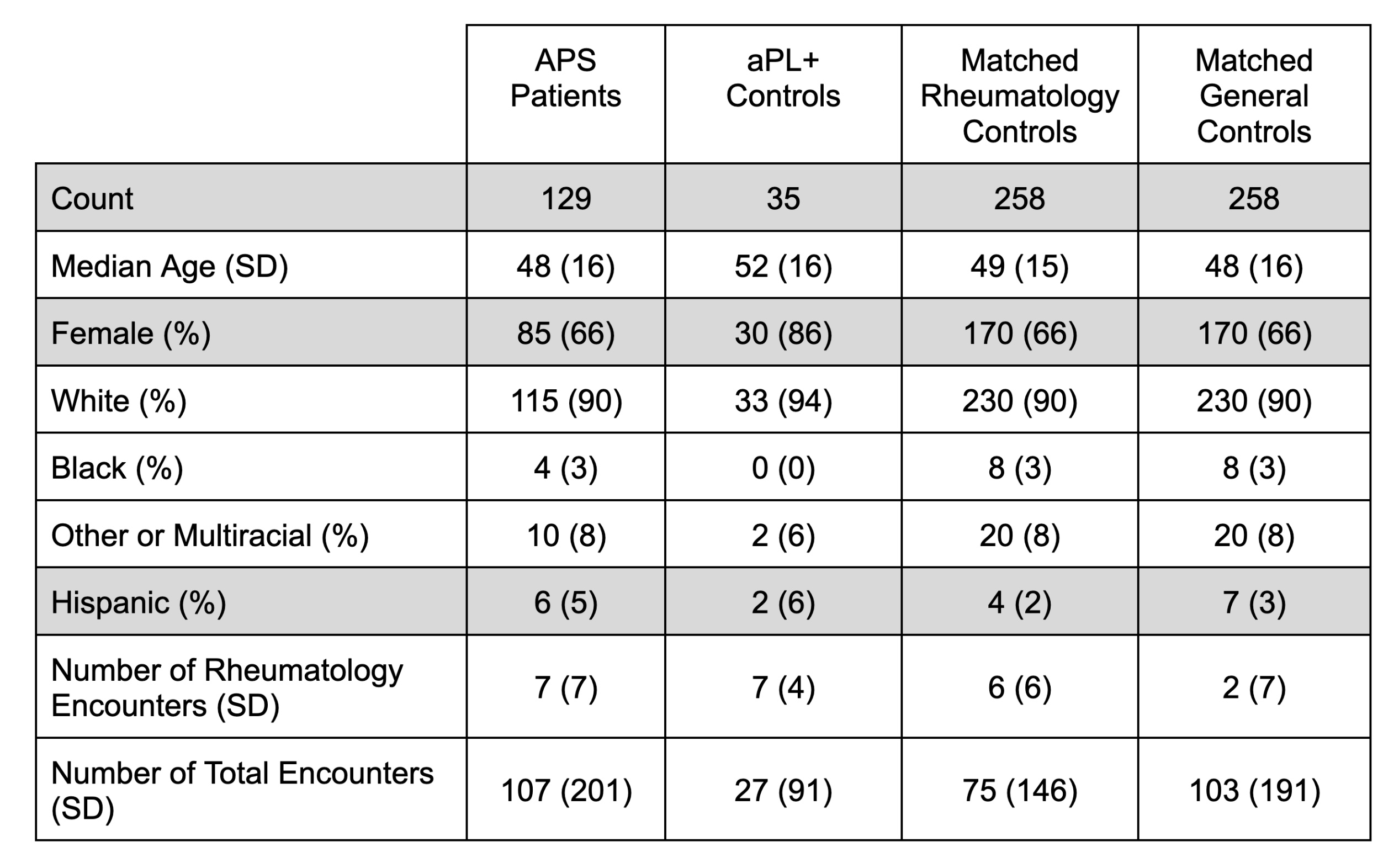
Methods: Data from a single United States-based academic medical center’s EHR (2015-2023) were used. The study population included 129 APS patients–classification manually verified by APS experts–and 2 control groups: 35 antiphospholipid antibody (aPL)-only patients (who had positive aPL tests, but did not meet the classification criteria for APS) and 258 controls (half with at least one rheumatology clinic visit and half without) matched for demographics and healthcare utilization (Table 1). Structured EHR data for ICD-10 codes, medications, and laboratory tests were engineered into 1,878 features. The recursive-partitioning (‘rpart’) R package (version 4.1.19) was trained to classify APS vs. all controls using a decision tree of depth 3, 4, and 5. These decision trees were inspected by hand and merged using expert input to produce one final decision tree that could be evaluated on a held-out test set.
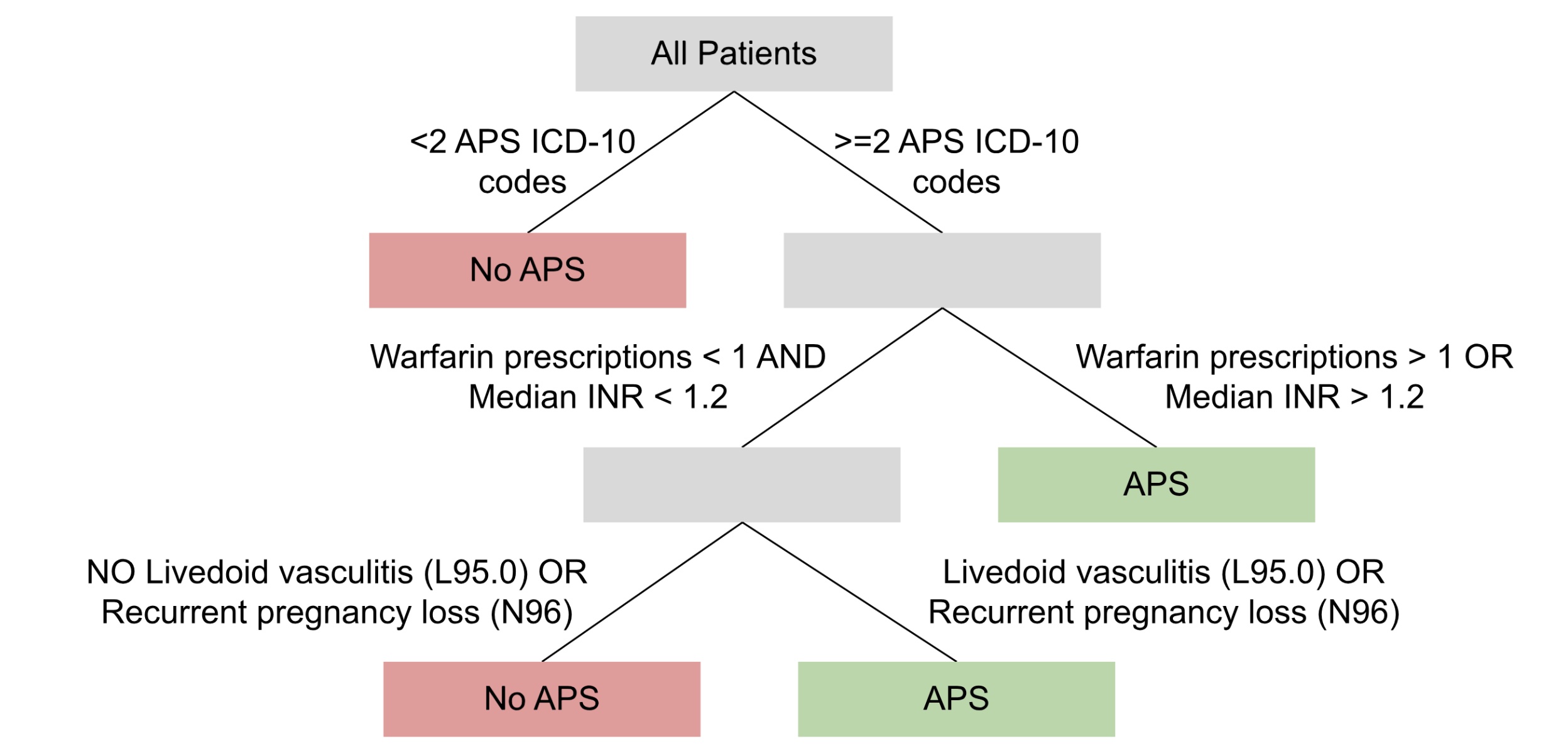
Results: The simplest possible rule-based computable phenotype for APS classified a patient as having APS if they had at least 1 diagnostic code for APS. This simple phenotype was perfectly sensitive (1.00) in our sample, but had only a moderate positive predictive value (PPV = 0.79) largely attributable to overcoding of APS diagnostic codes in aPL-only controls. Therefore, to identify APS patients with higher fidelity, we developed a decision tree using recursive partitioning and expert input (Figure 1). With the addition of the requirement for multiple APS diagnostic codes, medication usage, and some simple clinical features, the new model sacrificed sensitivity (0.78) for a much improved PPV (0.90). A potential limitation of this approach is that the false negatives had lower healthcare utilization (median = 19 encounters) than the true positives (median = 152), and therefore may be missing instrumental data required for correct classification.
Conclusion: This first computable phenotype lays a critical foundation for future APS research. The phenotype’s rule-based nature and relatively simple features should make it highly portable to other health systems. Furthermore, the strong PPV is likely to allow researchers to conduct clinical research on groups of highly-likely APS patients, instead of using unreliable diagnostic codes alone. In the future, we hope to validate this phenotype among diverse health systems in pursuit of continuing to improve its sensitivity and inclusivity while maintaining a high PPV.
To cite this abstract in AMA style:
Balczewski E, Ambati A, Liang W, Madison J, Zuo Y, Singh K, Knight J. Electronic Health Record Rule-Based Computable Phenotype of Antiphospholipid Syndrome [abstract]. Arthritis Rheumatol. 2024; 76 (suppl 9). https://acrabstracts.org/abstract/electronic-health-record-rule-based-computable-phenotype-of-antiphospholipid-syndrome/. Accessed .« Back to ACR Convergence 2024
ACR Meeting Abstracts - https://acrabstracts.org/abstract/electronic-health-record-rule-based-computable-phenotype-of-antiphospholipid-syndrome/